SuPPLementarY InFormatIon10.1038...Supplementary information for reagent chemicals Gln analogs,...
Transcript of SuPPLementarY InFormatIon10.1038...Supplementary information for reagent chemicals Gln analogs,...

w w w. n a t u r e . c o m / n a t u r e | 1
SuPPLementarY InFormatIondoi:10.1038/nature10656
1
Supplementary information
Supplementary information for reagent chemicals
Gln analogs, 6-diazo-5-oxonorleucine (DON) and azaserine (AZA), enter
catalytic centers of a number of glutamine-utilizing enzymes, such as
Gln:fructose-6-phosphate amidotransferase (GFAT), which is a rate-limiting
enzyme of the HBP, and Gln:phosphoribosyl amidotransferase, which is
involved in the biosynthesis of inosine monophosphate. Due to the strong
activity of the diazo group, while they are frequently used as as HBP
inhibitors, many cytotoxic side-effects are also known.
Supplementary figure legends
Supplementary figure S1. Proposed model for the function of
GlcNAcylated H2B in the O-GlcNAc signaling pathway.
Glucose is metabolized to UDP-GlcNAc through the HBP, which branches off
from the glycolytic pathway. Because OGT activity depends on the
intracellular concentration of UDP-GlcNAc, the level of GlcNAcylation of
H2B S112 therefore reflects the availability of extracellular glucose. H2B
S112 GlcNAc is targeted by the BRE1 complex on the C-terminal α-helix of
H2B, facilitating ubiquitination of H2B K120. Hence, these coordinated

SUPPLEMENTARY INFORMATION
2 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S1
OHOH
HO
OOH
CH2OH
MLL5
GlcNAc
GlcNAc
RNAPII
MeK4
Ub
Me
K4
K120K120
RAD6
BRE1
E1
OGTs
ExtracellularGlucose
S112S112GlcNAc
UbK120K120
S112S112
HO
OOH
CH2OH
NHCOCH3
O
UDP
UDP-GlcNAc Glucose
HBP
Metabolic genes etc.
H2BH3K4HKMT
1
Supplementary information
Supplementary information for reagent chemicals
Gln analogs, 6-diazo-5-oxonorleucine (DON) and azaserine (AZA), enter
catalytic centers of a number of glutamine-utilizing enzymes, such as
Gln:fructose-6-phosphate amidotransferase (GFAT), which is a rate-limiting
enzyme of the HBP, and Gln:phosphoribosyl amidotransferase, which is
involved in the biosynthesis of inosine monophosphate. Due to the strong
activity of the diazo group, while they are frequently used as as HBP
inhibitors, many cytotoxic side-effects are also known.
Supplementary figure legends
Supplementary figure S1. Proposed model for the function of
GlcNAcylated H2B in the O-GlcNAc signaling pathway.
Glucose is metabolized to UDP-GlcNAc through the HBP, which branches off
from the glycolytic pathway. Because OGT activity depends on the
intracellular concentration of UDP-GlcNAc, the level of GlcNAcylation of
H2B S112 therefore reflects the availability of extracellular glucose. H2B
S112 GlcNAc is targeted by the BRE1 complex on the C-terminal α-helix of
H2B, facilitating ubiquitination of H2B K120. Hence, these coordinated
2
modifications likely promote transcriptional initiation of some metabolic
genes, as shown in H3K4me.
Supplementary Figure S2. Proteomic analysis of GlcNAcylated proteins in
chromatin.
a, Purification of GlcNAcylated glycoproteins from chromatin. The levels of
GlcNAc in chromatin in HeLa cells which were cultured in DMEM with 1 g/L
(Low) or 4.5 g/L (High) glucose, were analyzed by immunoblots. b, c,
Purification with α-O-GlcNAc (RL2) antibody and proteomic analysis. The
bound proteins were eluted with GlcNAc-O-serine (#1 and #2) or acidic
conditions (#3), and were analyzed by Ag staining (b) and WB (c). In, input;
E, elution from WGA beads; R, residual WGA resin. d, The O-GlcNAc
glycoproteins, which were purified from the chromatin of HeLa cells as
shown in Figure 1 and as listed in Supplementary Table S1, were
functionally categorized with the DAVID bioinformatic database. e, The
Venn diagram was obtained from a comparison of the 284 identified
O-GlcNAc glycoproteins with the 689 proteins previously reported by Wang
et al 20.
Supplementary figure S3. CBB-staining analysis for the purification of
recombinant OGT.

w w w. n a t u r e . c o m / n a t u r e | 3
SUPPLEMENTARY INFORMATION RESEARCH
RNA processing
Translation
Transport
Nuclear pore
RNA binding
Chromatin assemble
Tubulin
Heat shock
Unknown
Cytoskeleton
Total identified : 284 proteinsAnalyzed : 257 proteinsUnknown : 27 proteins
19.019.0
27.627.6
18.318.34.34.3
14.414.4
3.13.15.15.1
3.93.91.61.6 2.72.7
Supplementary Figure S2
d e
Wang et al.(689)
Fujiki et al.(284)
581 108 176
a b c
H3H2BH2AH4
In WGA
E R
IgG1
#2#1 #3
α-GlcNAc (RL2)
#2#1 #3
IgGH
IgGL
In WG
A
IgG1
α-GlcN
Ac (R
L2)
WB: α-H2A
WB: α-H4
HeLa S3 cells
Low/High glucose (24 h)
Chromatin extract (0.5 g)
WGA lectin
kDa
150
100
75
50
Low High
WB: α-GlcNAc (RL2)
α-GlcNAc (RL2) affinity
LC-MS/MS
WB: α-H3
WB: α-H2B
Glc:
kDa
250
150
50
75
37
25
100
15
20
Glc High: Glc High
2
modifications likely promote transcriptional initiation of some metabolic
genes, as shown in H3K4me.
Supplementary Figure S2. Proteomic analysis of GlcNAcylated proteins in
chromatin.
a, Purification of GlcNAcylated glycoproteins from chromatin. The levels of
GlcNAc in chromatin in HeLa cells which were cultured in DMEM with 1 g/L
(Low) or 4.5 g/L (High) glucose, were analyzed by immunoblots. b, c,
Purification with α-O-GlcNAc (RL2) antibody and proteomic analysis. The
bound proteins were eluted with GlcNAc-O-serine (#1 and #2) or acidic
conditions (#3), and were analyzed by Ag staining (b) and WB (c). In, input;
E, elution from WGA beads; R, residual WGA resin. d, The O-GlcNAc
glycoproteins, which were purified from the chromatin of HeLa cells as
shown in Figure 1 and as listed in Supplementary Table S1, were
functionally categorized with the DAVID bioinformatic database. e, The
Venn diagram was obtained from a comparison of the 284 identified
O-GlcNAc glycoproteins with the 689 proteins previously reported by Wang
et al 20.
Supplementary figure S3. CBB-staining analysis for the purification of
recombinant OGT.

SUPPLEMENTARY INFORMATION
4 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
rec. OG
T
kDa
15010075
50
37
25
20
Supplementary Figure S3
2
modifications likely promote transcriptional initiation of some metabolic
genes, as shown in H3K4me.
Supplementary Figure S2. Proteomic analysis of GlcNAcylated proteins in
chromatin.
a, Purification of GlcNAcylated glycoproteins from chromatin. The levels of
GlcNAc in chromatin in HeLa cells which were cultured in DMEM with 1 g/L
(Low) or 4.5 g/L (High) glucose, were analyzed by immunoblots. b, c,
Purification with α-O-GlcNAc (RL2) antibody and proteomic analysis. The
bound proteins were eluted with GlcNAc-O-serine (#1 and #2) or acidic
conditions (#3), and were analyzed by Ag staining (b) and WB (c). In, input;
E, elution from WGA beads; R, residual WGA resin. d, The O-GlcNAc
glycoproteins, which were purified from the chromatin of HeLa cells as
shown in Figure 1 and as listed in Supplementary Table S1, were
functionally categorized with the DAVID bioinformatic database. e, The
Venn diagram was obtained from a comparison of the 284 identified
O-GlcNAc glycoproteins with the 689 proteins previously reported by Wang
et al 20.
Supplementary figure S3. CBB-staining analysis for the purification of
recombinant OGT.
2
modifications likely promote transcriptional initiation of some metabolic
genes, as shown in H3K4me.
Supplementary Figure S2. Proteomic analysis of GlcNAcylated proteins in
chromatin.
a, Purification of GlcNAcylated glycoproteins from chromatin. The levels of
GlcNAc in chromatin in HeLa cells which were cultured in DMEM with 1 g/L
(Low) or 4.5 g/L (High) glucose, were analyzed by immunoblots. b, c,
Purification with α-O-GlcNAc (RL2) antibody and proteomic analysis. The
bound proteins were eluted with GlcNAc-O-serine (#1 and #2) or acidic
conditions (#3), and were analyzed by Ag staining (b) and WB (c). In, input;
E, elution from WGA beads; R, residual WGA resin. d, The O-GlcNAc
glycoproteins, which were purified from the chromatin of HeLa cells as
shown in Figure 1 and as listed in Supplementary Table S1, were
functionally categorized with the DAVID bioinformatic database. e, The
Venn diagram was obtained from a comparison of the 284 identified
O-GlcNAc glycoproteins with the 689 proteins previously reported by Wang
et al 20.
Supplementary figure S3. CBB-staining analysis for the purification of
recombinant OGT.
3
FLAG-tagged OGT was expressed by baculovirus in Sf9 insect cells according
to the manufacturer’s instructions (Invitrogen), and was purified by
anti-FLAG immunoaffinity purification. Arrowheads point to the indicated
proteins.
Supplementary figure S4. In vitro analysis of GlcNAcylation of fly histone
octamers.
Core histone octamers (0.5 µg) derived from human HeLa cells and fly S2
cells were reacted with recombinant OGT (0.5 µg) and UDP-[3H-]GlcNAc (0.2
µCi) for 10 hrs at 37 ºC. The reactions were resolved by SDS PAGE, and
electro-transferred onto the membrane. The reacted histone octamers were
visualized by CBB staining, and subjected to autoradiography.
Supplementary figure S5. q-TOF analysis of in vitro GlcNAcylated H2B.
H2B (0.5 µg) was GlcNAcylated by OGT (0.1 µg) in vitro, and the reactants
were analyzed by q-TOF MS.
Supplementary figure S6. Mapping of O-GlcNAc sites of H2B by
ETD-MS/MS analysis.
In vitro GlcNAcylated H2B (0.5 µg) was digested with trypsin, and the
glycopeptides were partially enriched with lectin-conjugated magnetic beads.

w w w. n a t u r e . c o m / n a t u r e | 5
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S4
Fly
(-)
Hum
an
1 2 3
H3H2BH2AH4
H2BAuto
CBB
kDa
9
16
9
16
3
FLAG-tagged OGT was expressed by baculovirus in Sf9 insect cells according
to the manufacturer’s instructions (Invitrogen), and was purified by
anti-FLAG immunoaffinity purification. Arrowheads point to the indicated
proteins.
Supplementary figure S4. In vitro analysis of GlcNAcylation of fly histone
octamers.
Core histone octamers (0.5 µg) derived from human HeLa cells and fly S2
cells were reacted with recombinant OGT (0.5 µg) and UDP-[3H-]GlcNAc (0.2
µCi) for 10 hrs at 37 ºC. The reactions were resolved by SDS PAGE, and
electro-transferred onto the membrane. The reacted histone octamers were
visualized by CBB staining, and subjected to autoradiography.
Supplementary figure S5. q-TOF analysis of in vitro GlcNAcylated H2B.
H2B (0.5 µg) was GlcNAcylated by OGT (0.1 µg) in vitro, and the reactants
were analyzed by q-TOF MS.
Supplementary figure S6. Mapping of O-GlcNAc sites of H2B by
ETD-MS/MS analysis.
In vitro GlcNAcylated H2B (0.5 µg) was digested with trypsin, and the
glycopeptides were partially enriched with lectin-conjugated magnetic beads.

SUPPLEMENTARY INFORMATION
6 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
+203 Da
+203 Da
+203 Da
unmodified
monoGlcNAcylated
triGlcNAcylated
diGlcNAcylated
13000 13500 14000 1425013250 13750 14500 14750 m/z15000
Supplementary Figure S5
3
FLAG-tagged OGT was expressed by baculovirus in Sf9 insect cells according
to the manufacturer’s instructions (Invitrogen), and was purified by
anti-FLAG immunoaffinity purification. Arrowheads point to the indicated
proteins.
Supplementary figure S4. In vitro analysis of GlcNAcylation of fly histone
octamers.
Core histone octamers (0.5 µg) derived from human HeLa cells and fly S2
cells were reacted with recombinant OGT (0.5 µg) and UDP-[3H-]GlcNAc (0.2
µCi) for 10 hrs at 37 ºC. The reactions were resolved by SDS PAGE, and
electro-transferred onto the membrane. The reacted histone octamers were
visualized by CBB staining, and subjected to autoradiography.
Supplementary figure S5. q-TOF analysis of in vitro GlcNAcylated H2B.
H2B (0.5 µg) was GlcNAcylated by OGT (0.1 µg) in vitro, and the reactants
were analyzed by q-TOF MS.
Supplementary figure S6. Mapping of O-GlcNAc sites of H2B by
ETD-MS/MS analysis.
In vitro GlcNAcylated H2B (0.5 µg) was digested with trypsin, and the
glycopeptides were partially enriched with lectin-conjugated magnetic beads.

w w w. n a t u r e . c o m / n a t u r e | 7
SUPPLEMENTARY INFORMATION RESEARCH
Full scan
ETD-MS/MS
Supplementary Figure S6a
GlcNAc
GlcNAc GlcNAc
GlcNAc

SUPPLEMENTARY INFORMATION
8 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Full scan
ETD-MS/MS
Supplementary Figure S6b
GlcNAc
GlcNAc GlcNAc
GlcNAc

w w w. n a t u r e . c o m / n a t u r e | 9
SUPPLEMENTARY INFORMATION RESEARCH
Full scan
ETD-MS/MS
Supplementary Figure S6c
GlcNAc
GlcNAc GlcNAc
GlcNAc
3
FLAG-tagged OGT was expressed by baculovirus in Sf9 insect cells according
to the manufacturer’s instructions (Invitrogen), and was purified by
anti-FLAG immunoaffinity purification. Arrowheads point to the indicated
proteins.
Supplementary figure S4. In vitro analysis of GlcNAcylation of fly histone
octamers.
Core histone octamers (0.5 µg) derived from human HeLa cells and fly S2
cells were reacted with recombinant OGT (0.5 µg) and UDP-[3H-]GlcNAc (0.2
µCi) for 10 hrs at 37 ºC. The reactions were resolved by SDS PAGE, and
electro-transferred onto the membrane. The reacted histone octamers were
visualized by CBB staining, and subjected to autoradiography.
Supplementary figure S5. q-TOF analysis of in vitro GlcNAcylated H2B.
H2B (0.5 µg) was GlcNAcylated by OGT (0.1 µg) in vitro, and the reactants
were analyzed by q-TOF MS.
Supplementary figure S6. Mapping of O-GlcNAc sites of H2B by
ETD-MS/MS analysis.
In vitro GlcNAcylated H2B (0.5 µg) was digested with trypsin, and the
glycopeptides were partially enriched with lectin-conjugated magnetic beads.

SUPPLEMENTARY INFORMATION
1 0 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
4
The supernatants (first and third columns) and eluates (second and fourth
columns) were subject to LC-ETD-MS/MS. Note that the indicated amino
acid numbers are counted from the first methionine. a, ETD spectrum of
m/z 731.6080 (3+) identified the peptide, which was derived from 78 to 94
amino acids of H2B with a single GlcNAc moiety attached to serine 91. b,
ETD spectrum of m/z 588.0620 (4+) identified the peptide, which was derived
from 95 to 114 amino acids of H2B with a single GlcNAc moiety attached to
serine 112. c, ETD spectrum of m/z 491.9090 (3+) identified the peptide,
which was derived from 115 to 126 amino acids of H2B with a single GlcNAc
moiety attached to serine 123.
Supplementary figure S7. Scanning for GlcNAc sites with the H2B
peptide library.
a, The sequences of H2B peptide library are listed. b, Positions of H2B
peptides are illustrated (upper). The values were normalized against cognate
alanine-mutated control peptides.
Supplementary figure S8. MALDI-TOF MS analysis of the number of
O-GlcNAc moieties attached to the H2B peptide.
The H2B peptide 101-115 (a) or the 111-125 peptide (b) was used as a
substrate for the in vitro OGT assay. The reacted peptides (0.5 µg) were

w w w. n a t u r e . c o m / n a t u r e | 1 1
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S7
Covering H2B peptide control peptide(a.a.) (Original) (Ser/Thr to Ala)
1 -15 PDPAKSAPAPKKGSK PDPAKAAPAPKKGAK6 -20 SAPAPKKGSKKAVTK AAPAPKKGAKKAVAK11 -25 KKGSKKAVTKAQKKD KKGAKKAVAKAQKKD16 -30 KAVTKAQKKDGKERK KAVAKAQKKDGKERK21 -35 AQKKDGKERKRSRKE AQKKDGKERKRARKE26 -40 GKERKRSRKESYSIY GKERKRARKEAYAIY31 -45 RSRKESYSIYVYKVL RARKEAYAIYVYKVL36 -50 SYSIYVYKVLKQVHP AYAIYVYKVLKQVHP41 -55 VYKVLKQVHPDTGIS VYKVLKQVHPDAGIA46 -60 KQVHPDTGISSKAMG KQVHPDAGIAAKAMG51 -66 DTGISSKAMGIMNSF DAGIAAKAMGIMNAF56 -70 SKAMGIMNSFVNDIF AKAMGIMNAFVNDIF61 -75 IMNSFVNDIFERIAG IMNAFVNDIFERIAG66 -80 VNDIFERIAGEASRL VNDIFERIAGEAARL71 -85 ERIAGEASRLAHYNK ERIAGEAARLAHYNK76 -90 EASRLAHYNKRSTIT EAARLAHYNKRAAIA81 -95 AHYNKRSTITSREIQ AHYNKRAAIAAREIQ86 -100 RSTITSREIQTAVRL RAAIAAREIQAAVRL91 -105 SREIQTAVRLLLPGE AREIQAAVRLLLPGE96 -110 TAVRLLLPGELAKHA AAVRLLLPGELAKHA101-115 LLPGELAKHAVSEGT LLPGELAKHAVAEGA106-120 LAKHAVSEGTKAVTK LAKHAVAEGAKAVAK111-125 VSEGTKAVTKYTSSK VAEGAKAVAKYAAAK
ba
10 20 30 40 50 60 70 80 90 100 110 1203
2
1
0
H2B
a.a
α1 α2 α3 αCL1 L2
Fold
O-G
lcN
Acyl
atio
n(W
T/m
utan
t)
N-terminal tail
4
The supernatants (first and third columns) and eluates (second and fourth
columns) were subject to LC-ETD-MS/MS. Note that the indicated amino
acid numbers are counted from the first methionine. a, ETD spectrum of
m/z 731.6080 (3+) identified the peptide, which was derived from 78 to 94
amino acids of H2B with a single GlcNAc moiety attached to serine 91. b,
ETD spectrum of m/z 588.0620 (4+) identified the peptide, which was derived
from 95 to 114 amino acids of H2B with a single GlcNAc moiety attached to
serine 112. c, ETD spectrum of m/z 491.9090 (3+) identified the peptide,
which was derived from 115 to 126 amino acids of H2B with a single GlcNAc
moiety attached to serine 123.
Supplementary figure S7. Scanning for GlcNAc sites with the H2B
peptide library.
a, The sequences of H2B peptide library are listed. b, Positions of H2B
peptides are illustrated (upper). The values were normalized against cognate
alanine-mutated control peptides.
Supplementary figure S8. MALDI-TOF MS analysis of the number of
O-GlcNAc moieties attached to the H2B peptide.
The H2B peptide 101-115 (a) or the 111-125 peptide (b) was used as a
substrate for the in vitro OGT assay. The reacted peptides (0.5 µg) were

SUPPLEMENTARY INFORMATION
1 2 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S8
a
H2B peptide (101-115)
H2B peptide (101-115)
H2B peptide (111-125)
H2B peptide (111-125)
+ GlcNAc- H2O
(203 Da)
+ GlcNAc- H2O
(203 Da)
bUDP-GlcNAc (-)
UDP-GlcNAc (+)
UDP-GlcNAc (-)
UDP-GlcNAc (+)
4
The supernatants (first and third columns) and eluates (second and fourth
columns) were subject to LC-ETD-MS/MS. Note that the indicated amino
acid numbers are counted from the first methionine. a, ETD spectrum of
m/z 731.6080 (3+) identified the peptide, which was derived from 78 to 94
amino acids of H2B with a single GlcNAc moiety attached to serine 91. b,
ETD spectrum of m/z 588.0620 (4+) identified the peptide, which was derived
from 95 to 114 amino acids of H2B with a single GlcNAc moiety attached to
serine 112. c, ETD spectrum of m/z 491.9090 (3+) identified the peptide,
which was derived from 115 to 126 amino acids of H2B with a single GlcNAc
moiety attached to serine 123.
Supplementary figure S7. Scanning for GlcNAc sites with the H2B
peptide library.
a, The sequences of H2B peptide library are listed. b, Positions of H2B
peptides are illustrated (upper). The values were normalized against cognate
alanine-mutated control peptides.
Supplementary figure S8. MALDI-TOF MS analysis of the number of
O-GlcNAc moieties attached to the H2B peptide.
The H2B peptide 101-115 (a) or the 111-125 peptide (b) was used as a
substrate for the in vitro OGT assay. The reacted peptides (0.5 µg) were
5
deionized with a reverse-phase micro column, and subjected to MALDI-TOF
MS analysis.
Supplementary figure S9. In vitro OGT assay with point mutants in the
N-terminal tail of H2B.
a, Illustration of the recombinant H2B mutants used for the in vitro OGT
assay. b, c, In vitro OGT assay using recombinant Xenopus H2B lacking its
N-terminal tail (H2BΔN, b) or substituted with the indicated Ser/Thr to Ala
(c). Recombinant H2B mutants (0.5 µg) were reacted with UDP-[3H-]GlcNAc
(0.2 µCi) by recombinant OGT (0.5 µg), and subjected to autoradiography
(upper) and CBB staining (bottom).
Supplementary figure S10. Generation and validation of the α-H2B S112
GlcNAc monoclonal antibody.
a, The peptide used for immunization. b, c, Validation of α-H2B S112 GlcNAc
antibody with ELISA (b), and WB analysis with in vitro GlcNAcylated H2B
and H2A (0.5 µg each, c). d, Validation of α-H2B S112 GlcNAc antibody.
Chromatin lysates, in which H2B was twice depleted (upper), were
individually subjected to WB (middle), and ChIP on peak 4888 (bottom).
Error bars, means and s.d. (n = 3). Sequences of the primer sets used for
ChIP assay were as follows: 5’- AGA CAA CGG CAA CCG AAA AG -3’, and 5’-

w w w. n a t u r e . c o m / n a t u r e | 1 3
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S9
kDa
Auto
CBB
9
16
9
16
kDa
Auto
CBB16
16
10 20 30 40 50 60 70 80 90 100 110 120
H2B 4 125
H2BΔN 12527
a.a
1C
ont.
H2B
H2B
ΔN
H2B
H2B
ΔN
2 3 4 5 6
hH2B
OGT + - +- + +1 2 3 4
H2B
H2B
S6A
H2B
S14A
H2B
T21A
a
b c
H2B S6A 4 125S
H2B S14A 4 125S
H2B T21A 4 125T
5
deionized with a reverse-phase micro column, and subjected to MALDI-TOF
MS analysis.
Supplementary figure S9. In vitro OGT assay with point mutants in the
N-terminal tail of H2B.
a, Illustration of the recombinant H2B mutants used for the in vitro OGT
assay. b, c, In vitro OGT assay using recombinant Xenopus H2B lacking its
N-terminal tail (H2BΔN, b) or substituted with the indicated Ser/Thr to Ala
(c). Recombinant H2B mutants (0.5 µg) were reacted with UDP-[3H-]GlcNAc
(0.2 µCi) by recombinant OGT (0.5 µg), and subjected to autoradiography
(upper) and CBB staining (bottom).
Supplementary figure S10. Generation and validation of the α-H2B S112
GlcNAc monoclonal antibody.
a, The peptide used for immunization. b, c, Validation of α-H2B S112 GlcNAc
antibody with ELISA (b), and WB analysis with in vitro GlcNAcylated H2B
and H2A (0.5 µg each, c). d, Validation of α-H2B S112 GlcNAc antibody.
Chromatin lysates, in which H2B was twice depleted (upper), were
individually subjected to WB (middle), and ChIP on peak 4888 (bottom).
Error bars, means and s.d. (n = 3). Sequences of the primer sets used for
ChIP assay were as follows: 5’- AGA CAA CGG CAA CCG AAA AG -3’, and 5’-

SUPPLEMENTARY INFORMATION
1 4 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S10
C-KHAVSEGTK-CONH2
CH2
GlcNAc
O
a d
c
α-H2B-S112GlcNAc
α-H2B
- + - +OGT
kDa
16
16
16
16
H2B H2A
α-H2A
α-GlcNAc (RL2)
b
e
10002000
40008000
1600032000
64000128000
3.5
3
2.5
2
1.5
1
0.5
0
Opt
ic d
ensi
ty
CKHAVS(GlcNAc)EGTK-NH2
CKHAVSEGTK-NH2
Dilution factor
0.1 μg
0.05 μg
0.025 μg
0.0125 μg
7 8
α-H2B S112 GlcNAc
H2B-C (S112 GlcNAc)
H2B-NH2B-N (S36 GlcNAc)
H2A (T101 GlcNAc)
H2AH2B-C
GlcNAc Ser
Ser
1 2 3 4 5 6
α-H2B
α-H2B
IP: IgGIP: H2B
Input
1st depletion
2nd depletion
α-H2B S112 GlcNAc
Input1st depletion
2nd depletion
Input1st depletion
2nd depletion
% in
put
0.0
0.2
0.4
0.6
0.8
1.0
peak 4888negative control
5
deionized with a reverse-phase micro column, and subjected to MALDI-TOF
MS analysis.
Supplementary figure S9. In vitro OGT assay with point mutants in the
N-terminal tail of H2B.
a, Illustration of the recombinant H2B mutants used for the in vitro OGT
assay. b, c, In vitro OGT assay using recombinant Xenopus H2B lacking its
N-terminal tail (H2BΔN, b) or substituted with the indicated Ser/Thr to Ala
(c). Recombinant H2B mutants (0.5 µg) were reacted with UDP-[3H-]GlcNAc
(0.2 µCi) by recombinant OGT (0.5 µg), and subjected to autoradiography
(upper) and CBB staining (bottom).
Supplementary figure S10. Generation and validation of the α-H2B S112
GlcNAc monoclonal antibody.
a, The peptide used for immunization. b, c, Validation of α-H2B S112 GlcNAc
antibody with ELISA (b), and WB analysis with in vitro GlcNAcylated H2B
and H2A (0.5 µg each, c). d, Validation of α-H2B S112 GlcNAc antibody.
Chromatin lysates, in which H2B was twice depleted (upper), were
individually subjected to WB (middle), and ChIP on peak 4888 (bottom).
Error bars, means and s.d. (n = 3). Sequences of the primer sets used for
ChIP assay were as follows: 5’- AGA CAA CGG CAA CCG AAA AG -3’, and 5’-

w w w. n a t u r e . c o m / n a t u r e | 1 5
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S11
1.28 5.24
88.27 5.21
6.69 0.04
93.23 0.04
Glc depletion 0 h
Glc depletion 24 h
PIPI
Annexin V
Annexin V
6
CAT AGC CAT CCA ATC GAA CG -3’ (peak 4888); 5’- CCA CCC CTT CCA
AAA TGA TG -3’, and 5’- TAG AGA TGG GTG GCT TCC TTT G -3’ (80 kbp
upstream from peak 4888 as a negative control). e, Dot blot analysis of the
cross-reactivity of α-H2B S112 GlcNAc antibody. The indicated amounts of
the indicated peptides and amino acids were spotted on a PVDF membrane,
and blotted with α-H2B S112 GlcNAc antibody. The sequences of the used
peptides were as follows: KHAV-S/S(GlcNAc)-EGTK (H2B-C
unmodified/S112 GlcNAc), SRKE-S/S(GlcNAc)-YSVY (H2B-N
unmodified/S36 GlcNAc), LGKV-S/S(GlcNAc)-IAQG (H2A unmodified/T101
GlcNAc).
Supplementary figure S11. Glucose depletion of HeLa cells: the effect of
cell death.
The cells were cultured in glucose-free DMEM for 24 hrs, and then were
subjected to flow cytometry with annexin V and PI staining. The value in
each partition shows the percentage of the cell populations.
Supplementary figure S12. Glucose depletion of HeLa cells: the effect on
histone modifications.
HeLa cells were cultured in glucose-free DMEM for the indicated time. The
histones, prepared from the cells, were analyzed by WB with antibodies
6
CAT AGC CAT CCA ATC GAA CG -3’ (peak 4888); 5’- CCA CCC CTT CCA
AAA TGA TG -3’, and 5’- TAG AGA TGG GTG GCT TCC TTT G -3’ (80 kbp
upstream from peak 4888 as a negative control). e, Dot blot analysis of the
cross-reactivity of α-H2B S112 GlcNAc antibody. The indicated amounts of
the indicated peptides and amino acids were spotted on a PVDF membrane,
and blotted with α-H2B S112 GlcNAc antibody. The sequences of the used
peptides were as follows: KHAV-S/S(GlcNAc)-EGTK (H2B-C
unmodified/S112 GlcNAc), SRKE-S/S(GlcNAc)-YSVY (H2B-N
unmodified/S36 GlcNAc), LGKV-S/S(GlcNAc)-IAQG (H2A unmodified/T101
GlcNAc).
Supplementary figure S11. Glucose depletion of HeLa cells: the effect of
cell death.
The cells were cultured in glucose-free DMEM for 24 hrs, and then were
subjected to flow cytometry with annexin V and PI staining. The value in
each partition shows the percentage of the cell populations.
Supplementary figure S12. Glucose depletion of HeLa cells: the effect on
histone modifications.
HeLa cells were cultured in glucose-free DMEM for the indicated time. The
histones, prepared from the cells, were analyzed by WB with antibodies

SUPPLEMENTARY INFORMATION
1 6 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Glc depletion (h)
α-H3 K4me3
α-H3 K14Ac
α-H2B K120ub
α-H2B S112GlcNAc
36120 24 48
α-H2B
α-H3 K56Ac
α-H4 K16Ac
Supplementary Figure S12
6
CAT AGC CAT CCA ATC GAA CG -3’ (peak 4888); 5’- CCA CCC CTT CCA
AAA TGA TG -3’, and 5’- TAG AGA TGG GTG GCT TCC TTT G -3’ (80 kbp
upstream from peak 4888 as a negative control). e, Dot blot analysis of the
cross-reactivity of α-H2B S112 GlcNAc antibody. The indicated amounts of
the indicated peptides and amino acids were spotted on a PVDF membrane,
and blotted with α-H2B S112 GlcNAc antibody. The sequences of the used
peptides were as follows: KHAV-S/S(GlcNAc)-EGTK (H2B-C
unmodified/S112 GlcNAc), SRKE-S/S(GlcNAc)-YSVY (H2B-N
unmodified/S36 GlcNAc), LGKV-S/S(GlcNAc)-IAQG (H2A unmodified/T101
GlcNAc).
Supplementary figure S11. Glucose depletion of HeLa cells: the effect of
cell death.
The cells were cultured in glucose-free DMEM for 24 hrs, and then were
subjected to flow cytometry with annexin V and PI staining. The value in
each partition shows the percentage of the cell populations.
Supplementary figure S12. Glucose depletion of HeLa cells: the effect on
histone modifications.
HeLa cells were cultured in glucose-free DMEM for the indicated time. The
histones, prepared from the cells, were analyzed by WB with antibodies
7
against the indicated histone modifications.
Supplementary figure S13. The effect of retreatment with the physiologic
concentrations of glucose.
HeLa cells were cultured in glucose-free DMEM for 24 hrs. After glucose
depletion, the cells were further cultured in the media, which were
supplemented with the indicated concentrations of glucose, or 150 µM
PUGNAc for 24 hrs.
Supplementary figure S14. Generation and validation of retroviruses
delivering shRNA against OGT.
a, The sequence of shRNA against OGT (shOGT) and the structure of
recombinant retrovirus is shown (upper). The experimental procedure is
shown (middle). The shOGT was retrovirally transferred into HeLa cells,
and the infected cells were sorted as ZsGreen -positive cells. The sorting of
the infected cells was validated by flow cytometry (lower). b, The efficiency of
the OGT knockdown was confirmed by WB. The intensities of the bands were
quantified with the ImageJ program. c, The GlcNAc level in chromatin
derived from the cells knocked down with OGT.
Supplementary figure S15. Analysis of nucleosome incorporation of

w w w. n a t u r e . c o m / n a t u r e | 1 7
SUPPLEMENTARY INFORMATION RESEARCH
α-H2B S112GlcNAc
α-H2B
1.00.5 PUG0 4.5Glc (g/L)
Supplementary Figure S13
7
against the indicated histone modifications.
Supplementary figure S13. The effect of retreatment with the physiologic
concentrations of glucose.
HeLa cells were cultured in glucose-free DMEM for 24 hrs. After glucose
depletion, the cells were further cultured in the media, which were
supplemented with the indicated concentrations of glucose, or 150 µM
PUGNAc for 24 hrs.
Supplementary figure S14. Generation and validation of retroviruses
delivering shRNA against OGT.
a, The sequence of shRNA against OGT (shOGT) and the structure of
recombinant retrovirus is shown (upper). The experimental procedure is
shown (middle). The shOGT was retrovirally transferred into HeLa cells,
and the infected cells were sorted as ZsGreen -positive cells. The sorting of
the infected cells was validated by flow cytometry (lower). b, The efficiency of
the OGT knockdown was confirmed by WB. The intensities of the bands were
quantified with the ImageJ program. c, The GlcNAc level in chromatin
derived from the cells knocked down with OGT.
Supplementary figure S15. Analysis of nucleosome incorporation of

SUPPLEMENTARY INFORMATION
1 8 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S14
a b
Infection Sortingfor ZsGreen+ cells
shOGT/pSIREN-ZsGreen ZsGreenLTR shOGT LTR
shLuc :5'-TGC GTT GCT AGT ACC AAC-3'
shOGT :5'-GCA CAT AGC AAT CTG GCT TCC-3'
shLuc shOGT
kDa150
100
75
c
α-GlcNAc (RL2)kDa
GlcCont. GlcCont.
shLuc shOGT (1291)
26
16
38
α-H2B
α-OGT
α-βActin
1.0 0.15
ZsGreen
89.5 %
97.8 %
ZsGreen
10.1 %
97.8 %
7
against the indicated histone modifications.
Supplementary figure S13. The effect of retreatment with the physiologic
concentrations of glucose.
HeLa cells were cultured in glucose-free DMEM for 24 hrs. After glucose
depletion, the cells were further cultured in the media, which were
supplemented with the indicated concentrations of glucose, or 150 µM
PUGNAc for 24 hrs.
Supplementary figure S14. Generation and validation of retroviruses
delivering shRNA against OGT.
a, The sequence of shRNA against OGT (shOGT) and the structure of
recombinant retrovirus is shown (upper). The experimental procedure is
shown (middle). The shOGT was retrovirally transferred into HeLa cells,
and the infected cells were sorted as ZsGreen -positive cells. The sorting of
the infected cells was validated by flow cytometry (lower). b, The efficiency of
the OGT knockdown was confirmed by WB. The intensities of the bands were
quantified with the ImageJ program. c, The GlcNAc level in chromatin
derived from the cells knocked down with OGT.
Supplementary figure S15. Analysis of nucleosome incorporation of

w w w. n a t u r e . c o m / n a t u r e | 1 9
SUPPLEMENTARY INFORMATION RESEARCH
Flag: Cont.
S P S P S P S P
H2B(WT)
H2B(AA)
H2B(K120R)
Supplementary Figure S15
α-FLAG
α-H2B
EtBr200 b
p lad
der
7
against the indicated histone modifications.
Supplementary figure S13. The effect of retreatment with the physiologic
concentrations of glucose.
HeLa cells were cultured in glucose-free DMEM for 24 hrs. After glucose
depletion, the cells were further cultured in the media, which were
supplemented with the indicated concentrations of glucose, or 150 µM
PUGNAc for 24 hrs.
Supplementary figure S14. Generation and validation of retroviruses
delivering shRNA against OGT.
a, The sequence of shRNA against OGT (shOGT) and the structure of
recombinant retrovirus is shown (upper). The experimental procedure is
shown (middle). The shOGT was retrovirally transferred into HeLa cells,
and the infected cells were sorted as ZsGreen -positive cells. The sorting of
the infected cells was validated by flow cytometry (lower). b, The efficiency of
the OGT knockdown was confirmed by WB. The intensities of the bands were
quantified with the ImageJ program. c, The GlcNAc level in chromatin
derived from the cells knocked down with OGT.
Supplementary figure S15. Analysis of nucleosome incorporation of
8
exogenously expressed FLAG-tagged H2B mutants.
Non-chromatin proteins were extracted by 0.1% NP40 treatment of the HeLa
cells which were stably expressing FLAG-tagged H2B mutants (S;
supernatant). The histones in chromatin were solubilized by micrococcal
nuclease digestion of the residual pellet fractions (P; pellet).
Supplementary figure S16. Reagents affecting cellular glycolysis and
protein GlcNAcylation.
a, HeLa cells were cultured in glucose-free DMEM for 24 hrs, and then were
treated with 1 g/L D-glucose, 1 g/L L-glucose (not transported into cells), 1
g/L 3-O-methyl-D-glucopyranose (transported, but not phosphorylated at the
C-6 position), or 1 g/L 2-deoxyglucose (transported, phosphorylated at the
C-6 position, but not at the C-1 position) for an additional 24 hrs. The
chromatins derived from the treated cells were subject to WB with the
indicated antibodies. b, HeLa cells were cultured in DMEM in the presence
or the absence of PUGNAc (an inhibitor of OGA) and 0.1 mM iodoacetate (an
inhibitor of glyceraldehyde-3-phosphate dehydrogenase, GAPDH) for 24 hrs.
Of note, while iodoacetate is frequently used for inhibiting GAPDH in cells or
tissues, it often provokes a number of side effects that are caused by the
nonspecific reaction of iodoacetate with thiol reagents.

SUPPLEMENTARY INFORMATION
2 0 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S16
IAA
PUG
α-H2B K120ub
α-H2B S112GlcNAc
α-H2B
α-H2B K120ub
α-H2B S112GlcNAc
α-H2B
L-glucose
3-O-methyl-D-glucopyranose
2-deoxyglucose
ContD-glucose
a b
-
- -
+ +
+
8
exogenously expressed FLAG-tagged H2B mutants.
Non-chromatin proteins were extracted by 0.1% NP40 treatment of the HeLa
cells which were stably expressing FLAG-tagged H2B mutants (S;
supernatant). The histones in chromatin were solubilized by micrococcal
nuclease digestion of the residual pellet fractions (P; pellet).
Supplementary figure S16. Reagents affecting cellular glycolysis and
protein GlcNAcylation.
a, HeLa cells were cultured in glucose-free DMEM for 24 hrs, and then were
treated with 1 g/L D-glucose, 1 g/L L-glucose (not transported into cells), 1
g/L 3-O-methyl-D-glucopyranose (transported, but not phosphorylated at the
C-6 position), or 1 g/L 2-deoxyglucose (transported, phosphorylated at the
C-6 position, but not at the C-1 position) for an additional 24 hrs. The
chromatins derived from the treated cells were subject to WB with the
indicated antibodies. b, HeLa cells were cultured in DMEM in the presence
or the absence of PUGNAc (an inhibitor of OGA) and 0.1 mM iodoacetate (an
inhibitor of glyceraldehyde-3-phosphate dehydrogenase, GAPDH) for 24 hrs.
Of note, while iodoacetate is frequently used for inhibiting GAPDH in cells or
tissues, it often provokes a number of side effects that are caused by the
nonspecific reaction of iodoacetate with thiol reagents.

w w w. n a t u r e . c o m / n a t u r e | 2 1
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S17
α-BRE1
α-H2B S112 GlcNAc
IP: IgGIP: H2B S112 GlcNAc
a bα-BRE1
α-Flag (H2B)
IP:Flag-: Cont. H2B (WT)
Cont. Glc Cont. Glc
9
Supplementary figure S17. WB analysis of the glucose-dependent
interaction between H2B S112 GlcNAc and BRE1A.
a, FLAG-tagged H2B was expressed in HeLa cells which were cultured in
media with or without glucose. H2B was immunoprecipitated with
anti-FLAG antibody, and then subjected to WB analysis with α-BRE1A
antibody. b, H2B S112 GlcNAc was immunoprecipitated from HeLa cells
cultured under normal condition (i.e. glucose (+)-DMEM supplemented with
10 % FBS), and subjected to WB analysis with α-BRE1A antibody.
Supplementary figure S18. CBB staining analysis for the purification of
recombinant ubiquitin ligases of H2B.
a, CBB staining analysis of the purification of recombinant ubiquitin ligase
of H2B. Arrowheads point to the indicated proteins. b, In vitro
ubiquitination assay with H2B and recombinant E1-RAD6A-BRE1A/1B
ubiquitin ligases. H2B was ubiquitinated by the E1, RAD6A, and BRE1A/B
complex, and the reaction was subjected to WB analysis with α-H2B K120ub
antibody. The bands were quantified with ImageJ program (upper). Error
bars, means and s.d. (n = 3).
Supplementary figure S19. Analysis of sxc/ogt-mediated GlcNAcylation of
H2B in Drosophila polytene chromosomes.

SUPPLEMENTARY INFORMATION
2 2 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
rec. E1
rec. RA
D6A
rec. BR
E1 A
/1B com
plex
kDa
150
250
10075
50
37
2520
1015
150
250
100
75
50
37
2520
kDa
a b
Supplementary Figure S18
1 2 3 4- + + +E1+RAD6A+ - + ++BRE1A/1B
1 2 3 4Fold
(mon
o-ub
H2B
/H2B
)
0
1
2
3
n.d.
mono-ubH2B
H2B
di-ubH2Btri-ubH2B
**
α-H2B-K120ub
α-H2B
BRE1BBRE1A
9
Supplementary figure S17. WB analysis of the glucose-dependent
interaction between H2B S112 GlcNAc and BRE1A.
a, FLAG-tagged H2B was expressed in HeLa cells which were cultured in
media with or without glucose. H2B was immunoprecipitated with
anti-FLAG antibody, and then subjected to WB analysis with α-BRE1A
antibody. b, H2B S112 GlcNAc was immunoprecipitated from HeLa cells
cultured under normal condition (i.e. glucose (+)-DMEM supplemented with
10 % FBS), and subjected to WB analysis with α-BRE1A antibody.
Supplementary figure S18. CBB staining analysis for the purification of
recombinant ubiquitin ligases of H2B.
a, CBB staining analysis of the purification of recombinant ubiquitin ligase
of H2B. Arrowheads point to the indicated proteins. b, In vitro
ubiquitination assay with H2B and recombinant E1-RAD6A-BRE1A/1B
ubiquitin ligases. H2B was ubiquitinated by the E1, RAD6A, and BRE1A/B
complex, and the reaction was subjected to WB analysis with α-H2B K120ub
antibody. The bands were quantified with ImageJ program (upper). Error
bars, means and s.d. (n = 3).
Supplementary figure S19. Analysis of sxc/ogt-mediated GlcNAcylation of
H2B in Drosophila polytene chromosomes.

w w w. n a t u r e . c o m / n a t u r e | 2 3
SUPPLEMENTARY INFORMATION RESEARCH
+/+ Sxc1/Sxc7
Supplementary Figure S19
DAPI
H2B S112GlcNAc Merge
H3 K27 me3 DAPI
H2B S112GlcNAc Merge
H3 K27 me3
9
Supplementary figure S17. WB analysis of the glucose-dependent
interaction between H2B S112 GlcNAc and BRE1A.
a, FLAG-tagged H2B was expressed in HeLa cells which were cultured in
media with or without glucose. H2B was immunoprecipitated with
anti-FLAG antibody, and then subjected to WB analysis with α-BRE1A
antibody. b, H2B S112 GlcNAc was immunoprecipitated from HeLa cells
cultured under normal condition (i.e. glucose (+)-DMEM supplemented with
10 % FBS), and subjected to WB analysis with α-BRE1A antibody.
Supplementary figure S18. CBB staining analysis for the purification of
recombinant ubiquitin ligases of H2B.
a, CBB staining analysis of the purification of recombinant ubiquitin ligase
of H2B. Arrowheads point to the indicated proteins. b, In vitro
ubiquitination assay with H2B and recombinant E1-RAD6A-BRE1A/1B
ubiquitin ligases. H2B was ubiquitinated by the E1, RAD6A, and BRE1A/B
complex, and the reaction was subjected to WB analysis with α-H2B K120ub
antibody. The bands were quantified with ImageJ program (upper). Error
bars, means and s.d. (n = 3).
Supplementary figure S19. Analysis of sxc/ogt-mediated GlcNAcylation of
H2B in Drosophila polytene chromosomes.
10
The spread polytene chromosomes were prepared from wild-type flies (+/+)
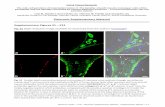
and OGT-null flies (sxc1/ sxc7), and were stained with α-H2B S112 GlcNAc
(green), α-H3K27me3 (red) and DAPI (blue).
Supplementary figure S20. Analysis of the cellular distribution of
GlcNAcylated H2B in HeLa cells.
HeLa cells were stained with α-H2B S112 GlcNAc fluorescent antibody
(green) and DAPI (white).
Supplementary figure S21. ChIP and WB analysis of the co-localization of
histone modifications.
Chromatin of HeLa cells was digested with MNase, and was
immunoprecipitated with the indicated antibodies. The native ChIP samples
were subjected to WB with H2B S112 GlcNAc monoclonal antibody or H2B
K120 ub antibody. Asterisk indicates nonspecific band.
Supplementary figure S22. Genome-wide analysis of distributions of H2B
S112 GlcNAc peaks.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B S112 GlcNAc peaks in the promoter, downstream and
other gene loci are shown with a comparison to the whole genome

SUPPLEMENTARY INFORMATION
2 4 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Supplementary Figure S20
DAPI Mergeα-H2B S112GlcNAc
10
The spread polytene chromosomes were prepared from wild-type flies (+/+)
and OGT-null flies (sxc1/ sxc7), and were stained with α-H2B S112 GlcNAc
(green), α-H3K27me3 (red) and DAPI (blue).
Supplementary figure S20. Analysis of the cellular distribution of
GlcNAcylated H2B in HeLa cells.
HeLa cells were stained with α-H2B S112 GlcNAc fluorescent antibody
(green) and DAPI (white).
Supplementary figure S21. ChIP and WB analysis of the co-localization of
histone modifications.
Chromatin of HeLa cells was digested with MNase, and was
immunoprecipitated with the indicated antibodies. The native ChIP samples
were subjected to WB with H2B S112 GlcNAc monoclonal antibody or H2B
K120 ub antibody. Asterisk indicates nonspecific band.
Supplementary figure S22. Genome-wide analysis of distributions of H2B
S112 GlcNAc peaks.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B S112 GlcNAc peaks in the promoter, downstream and
other gene loci are shown with a comparison to the whole genome

w w w. n a t u r e . c o m / n a t u r e | 2 5
SUPPLEMENTARY INFORMATION RESEARCH
WB:α-H2B S112GlcNAc
WB:α-H2B K120ub
H2B S112GlcNAc
H2B K120ub
H3 K9me2
H3 K27me3
H3 K4me2
IgG
*
Supplementary Figure S21
10
The spread polytene chromosomes were prepared from wild-type flies (+/+)
and OGT-null flies (sxc1/ sxc7), and were stained with α-H2B S112 GlcNAc
(green), α-H3K27me3 (red) and DAPI (blue).
Supplementary figure S20. Analysis of the cellular distribution of
GlcNAcylated H2B in HeLa cells.
HeLa cells were stained with α-H2B S112 GlcNAc fluorescent antibody
(green) and DAPI (white).
Supplementary figure S21. ChIP and WB analysis of the co-localization of
histone modifications.
Chromatin of HeLa cells was digested with MNase, and was
immunoprecipitated with the indicated antibodies. The native ChIP samples
were subjected to WB with H2B S112 GlcNAc monoclonal antibody or H2B
K120 ub antibody. Asterisk indicates nonspecific band.
Supplementary figure S22. Genome-wide analysis of distributions of H2B
S112 GlcNAc peaks.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B S112 GlcNAc peaks in the promoter, downstream and
other gene loci are shown with a comparison to the whole genome

SUPPLEMENTARY INFORMATION
2 6 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Promoter
<= 1000 bp <= 5000 bp <= 10000 bp
010
Per
cent
age
% 86
42
Genome
ChIP
0
Downstream
<= 1000 bp <= 5000 bp <= 10000 bp
10
Per
cent
age
% 86
42
Genome
ChIP
Gene
5’UTR 3’UTR Exon Intron All
Per
cent
age
%
8060
4020
0
Genome
ChIP
Supplementary Figure S22
10
The spread polytene chromosomes were prepared from wild-type flies (+/+)
and OGT-null flies (sxc1/ sxc7), and were stained with α-H2B S112 GlcNAc
(green), α-H3K27me3 (red) and DAPI (blue).
Supplementary figure S20. Analysis of the cellular distribution of
GlcNAcylated H2B in HeLa cells.
HeLa cells were stained with α-H2B S112 GlcNAc fluorescent antibody
(green) and DAPI (white).
Supplementary figure S21. ChIP and WB analysis of the co-localization of
histone modifications.
Chromatin of HeLa cells was digested with MNase, and was
immunoprecipitated with the indicated antibodies. The native ChIP samples
were subjected to WB with H2B S112 GlcNAc monoclonal antibody or H2B
K120 ub antibody. Asterisk indicates nonspecific band.
Supplementary figure S22. Genome-wide analysis of distributions of H2B
S112 GlcNAc peaks.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B S112 GlcNAc peaks in the promoter, downstream and
other gene loci are shown with a comparison to the whole genome
11
distribution. Analysis was performed on the web-based Cistrome analysis
pipeline module (http://cistrome.dfci.harvard.edu/).
Supplementary figure S23. H2B S112 GlcNAc was localized to the
transcribed genes.
a, The activity of the genes harboring H2B S112 GlcNAc in the promoter was
categorized into a high expression group (High) and a low expression group
(Low) based on the microarray datasets in Supplementary Table 2 [we
designated the ‘promoter’ as a region < 3 kbp upstream from the
transcription start site (TSS)]. b, The top five categories of gene ontology
analysis of biological processes of the genes harboring H2B S112 GlcNAc
peaks in the promoter are shown.
Supplementary figure S24. Genome-wide analysis of the H2B K120 ub
sites by ChIP-seq.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B K120 ub were averaged near TSS (left) and TTS (right)
(a). The average profiles of H2B S112 GlcNAc were overlaid. The distribution
of H2B K120 ub peaks in the promoter, downstream and other gene loci are
shown in comparison to the whole genome distribution (b). Analyses were
performed on the web-based Cistrome analysis pipeline module

w w w. n a t u r e . c o m / n a t u r e | 2 7
SUPPLEMENTARY INFORMATION RESEARCH
a
50 10075250 (%)
H2B S112 GlcNAc
High Low Expression level
76.0 24.0
Percentage
b
(%)
Cellular process
Metabolic process
Biological regulation
Primary metabolic process
Cellular metabolic process
80 60 40 20 0Percentage
1,134 (0.00180)
833 (0.00882)
808 (0.00283)
767 (0.00215)
741 (0.00114)Analyzed1,891 genes
Supplementary Figure S23
11
distribution. Analysis was performed on the web-based Cistrome analysis
pipeline module (http://cistrome.dfci.harvard.edu/).
Supplementary figure S23. H2B S112 GlcNAc was localized to the
transcribed genes.
a, The activity of the genes harboring H2B S112 GlcNAc in the promoter was
categorized into a high expression group (High) and a low expression group
(Low) based on the microarray datasets in Supplementary Table 2 [we
designated the ‘promoter’ as a region < 3 kbp upstream from the
transcription start site (TSS)]. b, The top five categories of gene ontology
analysis of biological processes of the genes harboring H2B S112 GlcNAc
peaks in the promoter are shown.
Supplementary figure S24. Genome-wide analysis of the H2B K120 ub
sites by ChIP-seq.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B K120 ub were averaged near TSS (left) and TTS (right)
(a). The average profiles of H2B S112 GlcNAc were overlaid. The distribution
of H2B K120 ub peaks in the promoter, downstream and other gene loci are
shown in comparison to the whole genome distribution (b). Analyses were
performed on the web-based Cistrome analysis pipeline module

SUPPLEMENTARY INFORMATION
2 8 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
Average Profile near TTS
0 1000 2000 3000-1000-2000-3000
1.15
1.10
1.05
1.00
0.95
0.90
Aver
age
prof
ile
Relative Distance to TTS (bp)
H2B K120 ubH2B S112 GlcNAc
0 1000 2000 3000-1000-2000-3000
1.30
1.25
1.20
1.15
1.10
1.05
Aver
age
prof
ile
Average Profile near TSS
Relative Distance to TSS (bp)
H2B K120 ubH2B S112 GlcNAc
Promoter
<= 1000 bp <= 5000 bp <= 10000 bp
010
Per
cent
age
%8
64
2
GenomeChIP
0
Downstream
<= 1000 bp <= 5000 bp <= 10000 bp
10P
erce
ntag
e %
86
42
GenomeChIP
Gene
5’UTR 3’UTR Exon Intron All
Per
cent
age
% 8060
4020
0
GenomeChIP
Supplementary Figure S24
a b
12
(http://cistrome.dfci.harvard.edu/).
Supplementary figure S25. ChIP-reChIP analysis of the co-localization of
H2B S112 GlcNAc and K120 ub.
a, The 3974 peaks co-localized with H2B S112 GlcNAc and K120 ub were
subdivided into five categories in terms of their overlapping length. b, The
top three peaks were picked from each category for a sequential ChIP
(α-H2B S112 GlcNAc antibody)-reChIP (α-H2B K120 ub antibody) analysis.
c, d, DNA library with ChIP (α-H2B S112 GlcNAc antibody, c) or reChIP
(α-H2B K120 ub antibody, d) was analyzed by qPCR. Error bars, means and
s.d. (n = 3). The sequences of the primers used for qPCR were as follows: 5’-
TTC AGG GGG AGA AAG CAA TG -3’ and 5’- TGC TGG GTA AAA GTG GAG
AGC -3’ (peak 4300); 5’- GAG AGA AGG CCT CCA AAA TCT C -3’ and 5’-
ACA TTG CCA CAT TGC CAC AG -3’ (peak 23556); 5’- TTT CTC AGC TTC
TGG CCA TC -3’ and 5’- AAT TTG AGT GGG GAC ACA GAG C -3’ (peak
817); 5’- AAG GAA AGG GAA AGG GAA GG -3’ and 5’- TTG CAT GCT GAT
GGT GAG TG -3’ (peak 25177); 5’- GCC CAT CTG ATT TGG GAA AC -3’ and
5’- ATA GGC ATG AAC CAC TGT GCT G -3’ (peak 42036); 5’- TCA TCC CAT
GAT GGC TTC AG -3’ and 5’- GAA GCA TTG GCA CTT TCT CG -3’ (peak
16699); 5’- ATT TCA ACC CAG CCA AGA GC -3’ and 5’- TGT GTC TTC CTC
CCA TGT TCT G -3’ (peak 28633); 5’- AGG CAG AAA TGG AAT CCT ACC C
11
distribution. Analysis was performed on the web-based Cistrome analysis
pipeline module (http://cistrome.dfci.harvard.edu/).
Supplementary figure S23. H2B S112 GlcNAc was localized to the
transcribed genes.
a, The activity of the genes harboring H2B S112 GlcNAc in the promoter was
categorized into a high expression group (High) and a low expression group
(Low) based on the microarray datasets in Supplementary Table 2 [we
designated the ‘promoter’ as a region < 3 kbp upstream from the
transcription start site (TSS)]. b, The top five categories of gene ontology
analysis of biological processes of the genes harboring H2B S112 GlcNAc
peaks in the promoter are shown.
Supplementary figure S24. Genome-wide analysis of the H2B K120 ub
sites by ChIP-seq.
The results from Cis-regulatory Element Annotation System (CEAS). The
distributions of H2B K120 ub were averaged near TSS (left) and TTS (right)
(a). The average profiles of H2B S112 GlcNAc were overlaid. The distribution
of H2B K120 ub peaks in the promoter, downstream and other gene loci are
shown in comparison to the whole genome distribution (b). Analyses were
performed on the web-based Cistrome analysis pipeline module

w w w. n a t u r e . c o m / n a t u r e | 2 9
SUPPLEMENTARY INFORMATION RESEARCH
Supplementary Figure S25
> 10 10-100 100-500 500-1000 < 1000
ChIP: α-H2B S112GlcNAc
% in
put
0.0
0.2
0.4
0.6
0.8
1.0
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 N
% C
hIP
: α-H
2B S
112G
lcN
Ac
0
2
4
6
8
10
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 N
ChIP: α-H2B S112GlcNAcRe-ChIP: α-H2B K120Ub
bp peak number> 10 1210 -100 167100-500 2105500-1000 1544 < 1000 146total 3974
1 peak 43002 peak 235563 peak 8174 peak 251775 peak 420366 peak 166997 peak 28633 8 peak 427489 peak 743610 peak 151411 peak 575612 peak 2862513 peak 488814 peak 487815 peak 44128
> 10 10-100 100-500 500-1000 < 1000
n.d.n.d.n.d. n.d.
a c
db
12
(http://cistrome.dfci.harvard.edu/).
Supplementary figure S25. ChIP-reChIP analysis of the co-localization of
H2B S112 GlcNAc and K120 ub.
a, The 3974 peaks co-localized with H2B S112 GlcNAc and K120 ub were
subdivided into five categories in terms of their overlapping length. b, The
top three peaks were picked from each category for a sequential ChIP
(α-H2B S112 GlcNAc antibody)-reChIP (α-H2B K120 ub antibody) analysis.
c, d, DNA library with ChIP (α-H2B S112 GlcNAc antibody, c) or reChIP
(α-H2B K120 ub antibody, d) was analyzed by qPCR. Error bars, means and
s.d. (n = 3). The sequences of the primers used for qPCR were as follows: 5’-
TTC AGG GGG AGA AAG CAA TG -3’ and 5’- TGC TGG GTA AAA GTG GAG
AGC -3’ (peak 4300); 5’- GAG AGA AGG CCT CCA AAA TCT C -3’ and 5’-
ACA TTG CCA CAT TGC CAC AG -3’ (peak 23556); 5’- TTT CTC AGC TTC
TGG CCA TC -3’ and 5’- AAT TTG AGT GGG GAC ACA GAG C -3’ (peak
817); 5’- AAG GAA AGG GAA AGG GAA GG -3’ and 5’- TTG CAT GCT GAT
GGT GAG TG -3’ (peak 25177); 5’- GCC CAT CTG ATT TGG GAA AC -3’ and
5’- ATA GGC ATG AAC CAC TGT GCT G -3’ (peak 42036); 5’- TCA TCC CAT
GAT GGC TTC AG -3’ and 5’- GAA GCA TTG GCA CTT TCT CG -3’ (peak
16699); 5’- ATT TCA ACC CAG CCA AGA GC -3’ and 5’- TGT GTC TTC CTC
CCA TGT TCT G -3’ (peak 28633); 5’- AGG CAG AAA TGG AAT CCT ACC C

SUPPLEMENTARY INFORMATION
3 0 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
13
-3’ and 5’- AAT GGG GAT GCA GAG GAA AC -3’ (peak 42728); 5’- TGA AAA
GGA TGC TGC GAG TG -3’ and 5’- TCC CTC TTT GCC ACT ATG CTC -3’
(peak 7436); 5’- TTT GAT CAG GGA CCC AAA CC -3’ and 5’- TTC CCT TTG
GGT TGG TCA AG -3’ (peak 1514); 5’- TAT GGA CGG CAT GAT GGA TG -3’
and 5’- ATG TGA GCA ATG TGG GAA GG-3’ (peak 5756); 5’- ACT GCT TGC
AGC AAA TGT GG -3’ and 5’- ACT GCT TGC AGC AAA TGT GG -3’ (peak
28625); 5’- AGA CAA CGG CAA CCG AAA AG -3’ and 5’- CAT AGC CAT CCA
ATC GAA CG -3’ (peak 4888); 5’- TTT TCT CCT CCC AGG CCT TTA G -3’
and 5’- TGG GGA AAA TGT CTC CAA GG -3’ (peak 4878); 5’- AAC GTT GGA
AAT CCC AGA GC -3’ and 5’- CCA CCT CAA AAC GCA ACA AG -3’ (peak
44128); 5’- TCA CCA TTA GGG ACC TTC TTG C-3’ and 5’- TTC TGG GAT
TGC CTT TCC TG -3’ (GAPDH promoter as a negative control),
Supplementary table S1. List of identified GlcNAcylated proteins in
chromatin.
a, b, The eluted fractions were independently subjected to trypsin digestion,
and LC-MS/MS analysis to identify GlcNAcylated proteins in chromatin (the
mixture of elution #1 and #2 (a), and elution #3 (b), shown in Figure 1b).
Peptides were analyzed by Bioworks3.3 (SEQUEST algorithm) against a
non-redundant NCBI protein database and results were filtered according to
peptide probabilities, SEQUEST X-corr (XC) scores, peptide charge state,
12
(http://cistrome.dfci.harvard.edu/).
Supplementary figure S25. ChIP-reChIP analysis of the co-localization of
H2B S112 GlcNAc and K120 ub.
a, The 3974 peaks co-localized with H2B S112 GlcNAc and K120 ub were
subdivided into five categories in terms of their overlapping length. b, The
top three peaks were picked from each category for a sequential ChIP
(α-H2B S112 GlcNAc antibody)-reChIP (α-H2B K120 ub antibody) analysis.
c, d, DNA library with ChIP (α-H2B S112 GlcNAc antibody, c) or reChIP
(α-H2B K120 ub antibody, d) was analyzed by qPCR. Error bars, means and
s.d. (n = 3). The sequences of the primers used for qPCR were as follows: 5’-
TTC AGG GGG AGA AAG CAA TG -3’ and 5’- TGC TGG GTA AAA GTG GAG
AGC -3’ (peak 4300); 5’- GAG AGA AGG CCT CCA AAA TCT C -3’ and 5’-
ACA TTG CCA CAT TGC CAC AG -3’ (peak 23556); 5’- TTT CTC AGC TTC
TGG CCA TC -3’ and 5’- AAT TTG AGT GGG GAC ACA GAG C -3’ (peak
817); 5’- AAG GAA AGG GAA AGG GAA GG -3’ and 5’- TTG CAT GCT GAT
GGT GAG TG -3’ (peak 25177); 5’- GCC CAT CTG ATT TGG GAA AC -3’ and
5’- ATA GGC ATG AAC CAC TGT GCT G -3’ (peak 42036); 5’- TCA TCC CAT
GAT GGC TTC AG -3’ and 5’- GAA GCA TTG GCA CTT TCT CG -3’ (peak
16699); 5’- ATT TCA ACC CAG CCA AGA GC -3’ and 5’- TGT GTC TTC CTC
CCA TGT TCT G -3’ (peak 28633); 5’- AGG CAG AAA TGG AAT CCT ACC C

w w w. n a t u r e . c o m / n a t u r e | 3 1
SUPPLEMENTARY INFORMATION RESEARCH
13
-3’ and 5’- AAT GGG GAT GCA GAG GAA AC -3’ (peak 42728); 5’- TGA AAA
GGA TGC TGC GAG TG -3’ and 5’- TCC CTC TTT GCC ACT ATG CTC -3’
(peak 7436); 5’- TTT GAT CAG GGA CCC AAA CC -3’ and 5’- TTC CCT TTG
GGT TGG TCA AG -3’ (peak 1514); 5’- TAT GGA CGG CAT GAT GGA TG -3’
and 5’- ATG TGA GCA ATG TGG GAA GG-3’ (peak 5756); 5’- ACT GCT TGC
AGC AAA TGT GG -3’ and 5’- ACT GCT TGC AGC AAA TGT GG -3’ (peak
28625); 5’- AGA CAA CGG CAA CCG AAA AG -3’ and 5’- CAT AGC CAT CCA
ATC GAA CG -3’ (peak 4888); 5’- TTT TCT CCT CCC AGG CCT TTA G -3’
and 5’- TGG GGA AAA TGT CTC CAA GG -3’ (peak 4878); 5’- AAC GTT GGA
AAT CCC AGA GC -3’ and 5’- CCA CCT CAA AAC GCA ACA AG -3’ (peak
44128); 5’- TCA CCA TTA GGG ACC TTC TTG C-3’ and 5’- TTC TGG GAT
TGC CTT TCC TG -3’ (GAPDH promoter as a negative control),
Supplementary table S1. List of identified GlcNAcylated proteins in
chromatin.
a, b, The eluted fractions were independently subjected to trypsin digestion,
and LC-MS/MS analysis to identify GlcNAcylated proteins in chromatin (the
mixture of elution #1 and #2 (a), and elution #3 (b), shown in Figure 1b).
Peptides were analyzed by Bioworks3.3 (SEQUEST algorithm) against a
non-redundant NCBI protein database and results were filtered according to
peptide probabilities, SEQUEST X-corr (XC) scores, peptide charge state,
14
and two or more unique peptide hits per protein to eliminate false or
low-quality identifications. Protein name, p-value, SEQUEST Full (Sf) score,
SEQUEST X-corr (XC) score, molecular weight, accession no., and number of
peptides are indicated for each. c, 257 proteins were successfully converted
from 284 identified genes, listed in (a) and (b), and with the DAVID
bioinformatic database.
Supplementary table S2. Microarray analysis of HeLa cells cultured under
specific experimental conditions.
Total RNA was prepared from HeLa cells, which were cultured in DMEM
supplemented with 10% FBS (FBS), or glucose-free DMEM for 24 hrs
[Glucose (-)], or 4.5 g/L glucose re-treatment for 24 hrs after a 24 hr glucose
depletion [Glucose (+)] (n = 3, in each culture condition). The labeled cRNA
samples, which were obtained according to the standard protocols, were then
hybridized to a human U133 plus 2.0 microarray (Affymetrix).
Supplementary table S3. List of genes harboring H2B S112 GlcNAc in the
promoter or 50 kbp within the gene body, and the expression levels.
a, d, Gene symbols, which have H2B S112 GlcNAc peaks, H2B K120 ub
peaks and both in the promoter regions (a) or the regions between 50 kbp
upstream from the TSS and 50 kbp downstream from the TTS of each gene

SUPPLEMENTARY INFORMATION
3 2 | w w w. n a t u r e . c o m / n a t u r e
RESEARCH
14
and two or more unique peptide hits per protein to eliminate false or
low-quality identifications. Protein name, p-value, SEQUEST Full (Sf) score,
SEQUEST X-corr (XC) score, molecular weight, accession no., and number of
peptides are indicated for each. c, 257 proteins were successfully converted
from 284 identified genes, listed in (a) and (b), and with the DAVID
bioinformatic database.
Supplementary table S2. Microarray analysis of HeLa cells cultured under
specific experimental conditions.
Total RNA was prepared from HeLa cells, which were cultured in DMEM
supplemented with 10% FBS (FBS), or glucose-free DMEM for 24 hrs
[Glucose (-)], or 4.5 g/L glucose re-treatment for 24 hrs after a 24 hr glucose
depletion [Glucose (+)] (n = 3, in each culture condition). The labeled cRNA
samples, which were obtained according to the standard protocols, were then
hybridized to a human U133 plus 2.0 microarray (Affymetrix).
Supplementary table S3. List of genes harboring H2B S112 GlcNAc in the
promoter or 50 kbp within the gene body, and the expression levels.
a, d, Gene symbols, which have H2B S112 GlcNAc peaks, H2B K120 ub
peaks and both in the promoter regions (a) or the regions between 50 kbp
upstream from the TSS and 50 kbp downstream from the TTS of each gene
15
(d), are listed. b, The expression signals of 1,702 genes out of 2,048 genes
listed in (a), which have H2B S112 GlcNAc peaks in the promoter, are shown.
In the analysis, 346 genes were omitted due to gene conversion processes, in
which official gene symbols were converted from the Affymetrix probe ID for
expression levels (Supplementary table S2) and from the RefSeq number of
the genes with H2B S112 GlcNAc in the promoter by using DAVID
Bioinformatics Resources (http://david.abcc.ncifcrf.gov/). c, The 1,891 genes
were successfully converted from 2,048 genes harboring H2B S112 GlcNAc in
the promoter listed in (a), and were categorized by Gene Ontology analysis of
biological processes.