Racing Bacterial Cells in Microfluidic Gradients
-
Upload
matthew-thornton -
Category
Documents
-
view
23 -
download
0
description
Transcript of Racing Bacterial Cells in Microfluidic Gradients

Racing Bacterial Cells in Microfluidic Gradients
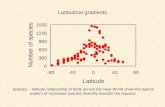
in order to measure chemotactic efficiency of isogenic bacteria population in correlation to their morphology

Racing Bacterial Cells in Microfluidic Gradients
in order to measure chemotactic efficiency of isogenic bacteria population in correlation to their morphology
Why:
Length variation is observed in isogenic bacteria population

Racing Bacterial Cells in Microfluidic Gradients
in order to measure chemotactic efficiency of isogenic bacteria population in correlation to their morphology
Why:
Length variation is observed in isogenic bacteria population
Does length variation have any functional role? → e.g. enhanced/diminshed motility?

Racing Bacterial Cells in Microfluidic Gradients
in order to measure chemotactic efficiency of isogenic bacteria population in correlation to their morphology
Why:
Length variation is observed in isogenic bacteria population
Does length variation have any functional role? → e.g. enhanced/diminshed motility?
Aim: Physical model of how cell size and number of flagella relate to swimming speeds and efficiency in chemotaxis

How:
Build microfluidics chamber using PDMS based soft-lithography

How:
Build microfluidics chamber using PDMS based soft-lithography Create nutrition gradient in chamber to induce chemotaxis (adding sugar)
→ Quantitative measurement of gradient by adding dye in same conc. → Simulating gradient with physics modeling program
chemoattractant bacteria

How:
Build microfluidics chamber using PDMS based soft-lithography Create nutrition gradient in chamber to induce chemotaxis (adding sugar)
→ Quantitative measurement of gradient by adding dye in same conc. → Simulating gradient with physics modeling program
Recording bacterias with DIC timelapse microscopy Identify single cells and measure their motion tracks (Matlab) as well as size
chemoattractant bacteria

How:
Build microfluidics chamber using PDMS based soft-lithography Create nutrition gradient in chamber to induce chemotaxis (adding sugar) → Quantitative measurement of gradient by adding dye in same conc.
→ Simulating gradient with physics modeling program Recording bacterias with DIC timelapse microscopy Identify single cells and measure their motion tracks (Matlab) as well as size

Build microfluidics chamber using PDMS based soft-lithography Create nutrition gradient in chamber to induce chemotaxis (adding sugar) → Quantitative measurement of gradient by adding dye in same conc.
→ Simulating gradient with physics modeling program Recording bacterias with DIC timelapse microscopy Identify single cells and measure their motion tracks (Matlab) as well as size
~1.5 µm thickness~ 3 µm spacing


Positions of tracked beads