The Central Dogma and Transcription Chapter 17: Sections 17.1-17.3.
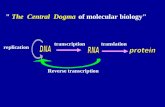
Chapter 21 (part 1) Transcription. Central Dogma.
-
date post
22-Dec-2015 -
Category
Documents
-
view
242 -
download
3
Transcript of Chapter 21 (part 1) Transcription. Central Dogma.

Chapter 21 (part 1)
Transcription

Central Dogma

Genes
• Sequence of DNA that is transcribed.• Encode proteins, tRNAs, rRNAs, etc..• “Housekeeping” genes encode
proteins or RNAs that are essential for normal cellular activity.
• Simplest bacterial genomes contain 500 to 600 genes.
• Mulitcellular Eukaryotes contain between 15,000 and 50,000 genes.

Types of RNAs
• tRNA, rRNA, and mRNA• rRNA and tRNA very abundant
relative to mRNA.• But mRNA is transcribed at
higher rates than rRNA and tRNA
• Abundance is a reflection of the relative stability of the different forms of RNA

RNA Content of E. coli Cells
typeSteady State Levels
Synthetic Capacity
Stability
rRNA 83% 58% High
tRNA 14% 10% High
mRNA 3% 32%Very Low

Phases of Transcription
• Initiation: Binding of RNA polymerase to promoter, unwinding of DNA, formation of primer.
• Elongation: RNA polymerase catalyzes the processive elongation of RNA chain, while unwinding and rewinding DNA strand
• Termination: termination of transcription and disassemble of transcription complex.

E. Coli RNA Polymerase• RNA polymerase core
enzyme is a multimeric protein ’
• The ’ subunit is involved in DNA binding
• The subunit contains the polymerase active site
• The subunit acts as scaffold on which the other subunits assemble.
• Also requires -factor for initiation –forms holo enzyme complex
Site of DNA binding and
RNA polymerization

-factor• The -factor is required for binding of the RNA
polymerase to the promoter
• Association of the RNA polynerase core complex w/ the -factor forms the holo-RNA polymerase complex
• W/o the -factor the core complex binds to DNA non-specifically.
• W/ the -factor, the holo-enzyme binds specifically with high affinity to the promoter region
• Also decreases the affinity of the RNA polymerase to non-promoter regions
• Different -factors for specific classes of genes

General Gene Structure
• Promoter – sequences recognized by RNA polymerase as start site for transcription.
• Transcribed region – template from which mRNA is synthesized
• Terminator – sequences signaling the release of the RNA polymerase from the gene.
5’ 3’Transcribed region terminatorPromoter

Gene Promoters• Site where RNA polymerase binds and
initiates transcription.• Gene that are regulated similarly contain
common DNA sequences (concensus sequences) within their promoters

Important Concensus Sequences
• Pribnow Box – position –10 from transcriptional start
• -35 region – position –35 from transcriptional start.
• Site where -factor binds.

Other -Factors
• Standard genes – 70
• Nitrogen regulated genes – 54
• Heat shock regulated genes – 32

How does RNA polymerase finds the
promoter?• RNA polymerase does not disassociate
from DNA strand and reassemble at the promoter (2nd order reaction – to slow)
• RNA polymerase holo-enzyme binds to DNA and scans for promoter sequences (scanning occurs in only one dimension, 100 times faster than diffusion limit)
• During scanning enzyme is bound non-specifically to DNA.
• Can quickly scan 2000 base pairs

Transcriptional Initiation
• Rate limiting step of trxn.• Requires unwinding of DNA and
synthesis of primer.• Conformational change occurs after DNA
binding of RNA polymerase holo-enzyme.• First RNA Polymerase binds to DNA
(closed-complex), then conformational change in the polymerase (open complex) causes formation of transcription bubble (strand separation).

Initiation of Polymerization
• RNA polymerase has two binding sites for NTPs
• Initiation site prefers to binds ATP and GTP (most RNAs begin with a purine at 5'-end)
• Elongation site binds the second incoming NTP
• 3'-OH of first attacks alpha-P of second to form a new phosphoester bond (eliminating PPi)
• When 6-10 unit oligonucleotide has been made, sigma subunit dissociates, completing "initiation“
• NusA protein binds to core complex after disassociation of -factor to convert RNA polymerase to elongation form.

Transcriptional Initiation
Closed complex
Open complex
Primer formation
Disassociation of -factor

Chain Elongation Core polymerase - no sigma
• Polymerase is accurate - only about 1 error in 10,000 bases
• Even this error rate is OK, since many transcripts are made from each gene
• Elongation rate is 20-50 bases per second - slower in G/C-rich regions (why??) and faster elsewhere
• Topoisomerases precede and follow polymerase to relieve supercoiling


Transcriptional Termination
• Process by which RNA polymerase complex disassembles from 3’ end of gene.
• Two Mechanisms – Pausing and “rho-mediated” termination

Pausing induces termination
• RNA polymerase can stall at “pause sites”
• Pause sites are GC rich (difficult to unwind)
• Can decrease trxn rates by a factor of 10 to 100.
• Hairpin formation in RNA can exaggerate pausing
• Hairpin structures in transcribed RNA can destabilize DNA:RNA hybrid in active site
• Nus A protein increases pausing when hairpins form.
3’end tends to be AU rich easily to disrupt during pausing. Leads to disassembly of RNA polymerase complex

Rho Dependent Termination
• rho is an ATP-dependent helicase
• it moves along RNA transcript, finds the "bubble", unwinds it and releases RNA chain


Eukaryotic Transcription
• Similar to what occurs in prokaryotes, but requires more accessory proteins in RNA polymerase complex.
• Multiple RNA polymerases

Eukaryotic RNA Polymerases
type Location Products
RNA polymerase I
Nucleolus rRNA
RNA polymerase II
Nucleoplasm
mRNA
RNA polymerase III
Nucleoplasm
rRNA, tRNA, others
Mitochondrial RNA polymerase
Mitochondria
Mitochondrial gene
transcripts
Chloroplast RNA polymerase
Chloroplast
Chloroplast gene
transcripts

Eukaryotic RNA Polymerases
• RNA polymerase I, II, and III
• All 3 are big, multimeric proteins (500-700 kD)
• All have 2 large subunits with sequences similar to and ' in E.coli RNA polymerase, so catalytic site may be conserved

Eukaryotic Gene Promoters• Contain AT rich concensus sequence
located –19 to –27 bp from transcription start (TATA box)
• Site where RNA polymerase II binds

RNA Polymerase II • Most interesting because it
regulates synthesis of mRNA • Yeast Pol II consists of 10 different
peptides (RPB1 - RPB10) • RPB1 and RPB2 are homologous to E.
coli RNA polymerase and ' • RPB1 has DNA-binding site; RPB2 binds
NTP • RPB1 has C-terminal domain (CTD) or
PTSPSYS • 5 of these 7 have -OH, so this is a
hydrophilic and phosphorylatable site

More RNA Polymerase II
• CTD is essential and this domain may project away from the globular portion of the enzyme (up to 50 nm!)
• Only RNA Pol II whose CTD is NOT phosphorylated can initiate transcription
• TATA box (TATAAA) is a consensus promoter
• 7 general transcription factors are required

Transcription Factors • Polymerase I, II, and III do not bind
specifically to promoters• They must interact with their
promoters via so-called transcription factors
• Transcription factors recognize and initiate transcription at specific promoter sequences

Transcription Factors• TFAIIA, TFAIIB –
components of RNA polymerase II holo-enzyme complex
• TFIID – Initiation factor, contains TATA binding protein (TBP) subunit. TATA box recognition.
• TFIIF – (RAP30/74) decrease affinity to non-promoter DNA

Eukaryotic Transcription
• Once initiation complex assembles process similar to bacteria (closed complex to open complex transition, primer formation)
• Once elongation phase begins most transcription factor disassociate from DNA and RNA polymerase II (but TFIIF may remain bound).
• TFIIS – Elongation factor binds at elongation phase. May also play analogous role to NusA protein in termination.