Zachary Neal Donnell A DISSERTATION PRESENTED TO ......at LCD, AphA is produced and LuxR is...
Transcript of Zachary Neal Donnell A DISSERTATION PRESENTED TO ......at LCD, AphA is produced and LuxR is...

REGULATION OF CQSA AND CQSS IN VIBRIO CHOLERAE
Zachary Neal Donnell
A DISSERTATION
PRESENTED TO THE FACULTY
OF PRINCETON UNIVERSITY
IN CANDIDACY FOR THE DEGREE
OF DOCTOR OF PHILOSOPHY
RECOMMENDED FOR ACCEPTANCE
BY THE DEPARTMENT OF
MOLECULAR BIOLOGY
[Advisor: Bonnie L. Bassler]
March 2015

ii
© Copyright by Zach Donnell, 2015. All rights reserved.

iii
TABLE OF CONTENTS Page number Abstract v Acknowledgements vi Chapter 1: An introduction to quorum sensing 1 Overview of quorum sensing 2
Quorum sensing in Gram-negative bacteria 3
Quorum sensing in Gram-positive bacteria 4
Quorum sensing in Vibrios 5
Feedback in V. cholerae quorum sensing 7
Chapter 2: Regulation of QS synthases and receptors in V. cholerae 16 Introduction 17 Results 17 Discussion 20 Materials and Methods 27 Chapter 3: Identification of regulators of CqsA and CqsS in V. cholerae 34 Introduction 35 Results 35 Discussion 38 Materials and Methods 47 Chapter 4: cqsA and cqsS regulation is post-transcriptional 57 Introduction 58 Results 58

iv
Discussion 62 Materials and Methods 71 Chapter 5: Examination of cqsA growth phase regulation 81 Introduction 82 Results 82 Discussion 86 Materials and Methods 93 References 100

v
Abstract Quorum sensing (QS) is a cell-to-cell communication mechanism used by
bacteria to determine their population density and to regulate genes involved in group
behaviors. The human pathogen Vibrio cholerae contains two distinct QS systems that
converge into a shared phosphorelay pathway. The cholera-specific pathway is
composed of the autoinducer CAI-1, which is produced by the synthase CqsA and
recognized by the membrane-bound histidine kinase CqsS. The universal QS pathway is
composed of the autoinducer AI-2, which is produced by the autoinducer synthase LuxS
and binds to the receptor LuxPQ. Here, we investigate regulation of the V. cholerae
autoiducer synthases and receptors. We discovered that the LuxS/PQ system exhibits
no regulation under conditions examined, whereas the CqsA/S system contains an auto-
regulatory positive feedback loop. We characterized feedback in the CqsA/S by
identifying the Qrr sRNAs as the source of feedback, and that they act via indirect
repression of CqsA/S through an unknown intermediate. Additionally, we showed that
regulation of cqsA and cqsS occurs at the post-transcriptional level. We further
investigated growth phase regulation of cqsA, and discovered DctD as a potential
regulator of the Qrr sRNAs.

vi
Acknowledgements
I would like to thank my wife Jiajia Ren and the Donnell family for emotional support, my
advisor Bonnie Bassler and the Bassler lab for scientific support, and the National
Science Foundation and the National Institutes of Health for funding support.

1
CHAPTER 1:
AN INTRODUCTION TO QUORUM SENSING

2
Overview of quorum sensing
Quorum sensing (QS) is a cell-to-cell communication mechanism in which
bacteria produce, release, and detect chemical messages to determine their population
density (Ng and Bassler, 2009). Information regarding population size drives bacterial
group behaviors because it allows bacteria to act as individuals or as collectives. Many
behaviors bacteria engage in require group participation to be effective, and consistent
with this notion, QS regulates processes including virulence, biofilm formation, and
bioluminescence (Ng and Bassler, 2009; Waters and Bassler, 2005).
The specific chemical messages bacteria use are called autoinducers (Nealson
and Hastings, 1979). Autoinducers range in structure from small molecules to
oligopeptides, and generally serve as signals specific to a particular species.
Autoinducer concentration serves as a proxy for the bacterial population size, and
detection of autoinducers controls the transition from individual to group behaviors. A
group of bacteria are in a low cell density (LCD) state when autoinducer concentration is
low and the bacteria exhibit individualistic behaviors (e.g. prioritizing cell growth over
production of bioluminescence). A group of bacteria are in a high cell density (HCD)
state when autoinducer concentrations have reached sufficient thresholds to induce
changes in gene expression that favor group behaviors.
Bacterial QS systems fall into two broad categories: Gram-negative QS systems
and Gram-positive QS systems. Gram-negative QS systems generally use small
signaling molecules and cytoplasmic or membrane-bound receptors, whereas Gram-
positive QS systems generally use peptides as the messages coupled with membrane-
bound receptors. In addition to these two primary categories, the QS systems of many
Vibrios contain features of both Gram-negative and Gram-positive QS systems,
specifically small signaling molecules coupled with membrane-bound receptors.

3
Quorum sensing in Gram-negative bacteria
The first QS system discovered was found in the marine bacterium Vibrio fischeri,
a symbiont of the bobtail squid Euprymna scolopes (Nealson et al., 1970). Initial studies
of this bacterium concerned the exclusive high cell density production of
bioluminescence via the luciferase operon (Figure 1.2). Two proteins, LuxI and LuxR,
were found to be responsible for controlling when the luciferase operon in V. fischeri was
expressed (Engebrecht and Silverman, 1984). LuxI, the autoinducer synthase, catalyzes
the production of the autoinducer N-3-(oxo-hexanoyl)-homoserine lactone (More et al.,
1996). The autoinducer acts as the ligand that activates LuxR, the master regulator of
the V. fischeri QS circuit (Engebrecht et al., 1983; Engebrecht and Silverman, 1984), and
serves to stabilize the intrinsically unstable protein. In addition to activating the genes
necessary for luciferase production, LuxR also activates the transcription of luxI, which
generates increased autoinducer. This positive feedback loop accelerates V. fischeri’s
transition from the LCD to HCD state (Nealson and Hastings, 1979).
Subsequent to the discovery of LuxI-R QS in V. fischeri, numerous other bacterial
species were shown to contain similar QS systems. For example, the QS circuit of the
opportunistic pathogen Pseudomonas aeruginosa contains two connected LuxI-R-type
QS systems (Passador et al., 1993; Pearson et al., 1994; Pearson et al., 1995). These
systems, LasI-R and RhlI-R, act in series to activate and repress the expression of
hundreds of genes in the P. aeruginosa genome, including genes necessary for toxin
production (Figure 1.3). In this case, the LasI-R system behaves like the LuxI-R system
in V. fischeri, except in addition to activating the LasI autoinducer synthase, LasR also
activates the RhlI synthase, thereby initiating a second LuxI-R type QS system (Pesci et
al., 1997). In addition to the LuxI-R type QS systems, P. aeruginosa also produces a

4
third chemical signal, 2-heptyl-3-hydroxy-4-quinolone (PQS), which further links the two
canonical QS circuits (Pesci et al., 1999).
Quorum sensing in Gram-positive bacteria
Gram-positive bacteria employ oligopeptides as their autoinducers. Oligopeptides
are transcribed and translated from genes and then are processed post-translationally,
often though circularization of the peptide backbone. Gram-positive bacteria use
membrane-bound histidine-kinases to detect autoinducer concentrations. Unlike
homoserine lactone autoinducers that diffuse through the lipid bilayer, oligopeptide
autoinducers must be actively transported across the membrane via membrane-bound
transporters.
The most thoroughly studied Gram-positive QS system is that of Staphylococcus
aureus (Havarstein et al., 1995; Ji et al., 1995). The primary components of the S.
aureus QS system are encoded in the agrBDCA operon (Figure 1.4). AgrB is a
membrane-bound protein responsible for processing the autoinducer precursor AgrD by
truncation and circularization into an autoinducing peptide (AIP), and then transporting
the signal oligopeptide across the lipid bilayer. When the extracellular AIP concentration
reaches a critical threshold, it binds to the receptor AgrC. AgrC and AgrA act as a two-
component system, in which AgrC is the membrane-bound histidine-kinase that passes
phosphate to the response regulator, AgrA. AgrA activates the transcription of the
divergent promoters P2 and P3.
In addition to the species-specific autoinducer signals, a unique autoinducer has
also been demonstrated to confer cell density information between bacterial species in
both Gram-negative and Gram-positive bacteria. Autoinducer 2 (AI-2) is a collection of
molecules that share the precursor DPD, 4,5-dihydroxy-2,3-pentanedione (Schauder et

5
al., 2001). DPD rapidly converts between alternative configurations, and some
configurations incorporate boron into the structure, as seen in V. harveyi AI-2 (Chen et
al., 2002). AI-2 is an extremely important aspect of QS because it reveals that QS-
communication can occur across species, and the population size of one bacterial
species can affect the gene expression of another.
Quorum sensing in Vibrios
The V. harveyi QS system is a composite of the canonical Gram-positive and
Gram-negative QS systems (Ng and Bassler, 2009; Waters and Bassler, 2005). The V.
harveyi system uses small-molecule autoinducers common to Gram-negative QS
systems, but these small molecules are detected by membrane bound receptors
common to Gram-positive QS systems. The V. harveyi system also uses three parallel
QS pathways that converge through a shared phosphorelay pathway to regulate
downstream gene expression.
The first autoinducer synthase in the system, LuxM (Bassler et al., 1993; Bassler
et al., 1994a), catalyzes the production of V. harveyi autoinducer 1 (HAI-1; N-(3-
hydroxybutyryl)-homoserine lactone) (Cao and Meighen, 1989). This autoinducer is
detected by the membrane-bound receptor LuxN (Freeman et al., 2000). The second
autoinducer synthase, CqsA (Miller et al., 2002), catalyzes the production of the so-
called cholera autoinducer 1 (CAI-1; (S)-3-hydroxytridecan-4-one) (Higgins et al., 2007),
and this autoinducer is detected by the membrane-bound receptor CqsS (Miller et al.
2002). The third autoinducer synthase in this system is LuxS, which catalyzes the
production of autoinducer 2 (AI-2; (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran
borate), and this autoinducer is detected by the membrane-bound receptor complex
LuxPQ (Neiditch et al., 2005; Neiditch et al., 2006). At LCD, LuxN, CqsS, and LuxPQ act

6
as kinases and funnel phosphate to the phosphorelay protein LuxU, which serves as the
point in the QS circuit where HAI-1, CAI-1, and AI-2 information converges (Henke and
Bassler, 2004; Miller et al., 2002). The phosphorelay protein LuxU passes phosphate to
the response regulator LuxO (Bassler et al., 1994b; Freeman and Bassler, 1999), which
binds DNA and activates the transcription of genes encoding five small RNAs (sRNAs)
termed quorum regulatory RNAs 1-5 (Qrr1-5) (Lenz et al., 2004). Qrr1-5 repress
translation of the QS master regulator LuxR by binding to and promoting degradation of
luxR mRNA (Tu and Bassler, 2007). Additionally, Qrr1-5 activate the expression of the
QS master regulator AphA by binding to the aphA transcript and inhibiting the formation
of an endogenous stem-loop that blocks AphA translation (Rutherford et al., 2011). Thus,
at LCD, AphA is produced and LuxR is repressed.
At high cell density, when autoinducers have accumulated, the autoinducers HAI-
1, CAI-1, and AI-2 bind their cognate receptors LuxN, CqsS, and LuxPQ, respectively. In
the bound state, these receptors act as phosphotases and reverse the flow of phosphate
through the phosphorelay. When LuxO is unphosphorylated, it does not activate the
transcription of qrr1-5. In the absence of Qrr1-5, luxR mRNA is translated, and LuxR
activates and represses hundreds of genes in the V. harveyi genome. Additionally, at
HCD when no Qrr sRNAs are present, aphA mRNA cannot be translated, and so no
AphA is made.
The Vibrio cholerae QS circuit is similar to that of V. harveyi, although there are a
few key differences. First, V. cholerae only possesses two parallel QS circuits (CqsA-
CqsS and LuxS-LuxPQ) as opposed to the three found in V. harveyi (Miller et al., 2002).
Second, V. cholerae contains only four Qrr sRNAs (Qrr1-4) unlike the five Qrr sRNAs in
V. harveyi. Third, Qrr1-4 in V. cholerae act redundantly to repress expression of hapR,

7
whereas Qrr1-5 in V. harveyi act additively to repress expression luxR. HapR and LuxR
are homologs.
Feedback in V. cholerae quorum sensing
Feedback plays a critical role in the robustness of information processing
pathways in V. cholerae. In addition to the core circuit of the V. cholerae QS system
described above, numerous feedback loops operate to optimize signal transmission
through the system. Here, the most important components of feedback in the V. cholerae
QS system are described.
Auto-repression occurs when a transcriptional regulator inhibits transcription of its
own RNA (Becskei and Serrano, 2000). Such feedback loops serve dual roles. First, the
feedback provides an upper bound on the production of the regulator. When regulator
levels reach a threshold determined by the strength of promoter binding, the regulators
bind their own promoter and occlude RNA polymerase from binding. Therefore, these
regulators turn off their own production only when a sufficient amount is already present
in a cell, reducing the dynamic range (Nevozhay et al., 2009). Second, auto-repression
dampens the noise between cells, meaning that the variation in regulator number
between cells is reduced (Paulsson, 2004). Ultimately, through the combination of these
two features, auto-repression loops serve to finely tune the concentration of the regulator
within and between cells.
In the V. cholerae QS system, three transcription factors are regulated via auto-
repression: LuxO, HapR, and AphA. LuxO, the activator of the Qrr sRNA genes, is an
NtrC-type activator, which bind DNA regardless of their phosphorylation state and
activate transcription though an ATP-driven mechanism. Although transcription factors in
this class typically act solely as activators, binding of the luxO promoter by LuxO

8
represses transcription and serves to repress luxO expression because binding does not
depend on phosphorylation (Tu et al., 2010). AphA and HapR, the master regulators of
the LCD and HCD responses, respectively, are capable of activating and repressing
promoters (Rutherford et al., 2011; Svenningsen et al., 2009). Both AphA and HapR bind
their own promoters and finely tune their expression through auto-repression.
Positive feedback also exists within the V. cholerae QS circuit. HapR activates
the expression of the qrr1-4 promoters indirectly through an unknown mechanism
(Svenningsen et al., 2009), and this architecture is important for the following reasons.
First, because Qrr1-4 are responsible for degrading the hapR mRNA, HapR activation of
qrr1-4 can serve to provide another check on maximum HapR levels. Furthermore, this
circuit architecture inherently accelerates V. cholerae’s transition from the HCD to LCD
state by increasing Qrr1-4 levels upon entry into an environment lacking autoinducers.
Negative feedback occurs within the circuit as well. First, similar to HapR
activation of qrr1-4, AphA represses the expression of these same sRNA genes. AphA is
expressed most highly at LCD, when Qrr1-4 are also most highly expressed. AphA
repression of qrr1-4 serves as a check on Qrr1-4 levels. If too much Qrr1-4 are produced
in a cell, AphA levels are increased and AphA repression of qrr1-4 is enhanced, which
then lowers Qrr1-4 concentration and AphA levels. Finally, AphA and HapR have also
been shown to repress the expression of one another (Rutherford et al., 2011). This
feature enhances the differences in gene expression between LCD and HCD states. At
LCD, AphA is expressed and represses HapR. When cells begin the transition from LCD
to HCD, HapR is produced and this boosts repression of AphA. Therefore, the system
set-up promotes commitment in master regulator expression.
Finally, as mentioned above, positive feedback between QS downstream
regulators and autoinducer production (autoinduction) has been shown to be a nearly

9
universal feature of QS systems in bacteria (Ng and Bassler, 2009; Shadel and Baldwin,
1991). However, no such mechanism has been demonstrated in V. cholerae. The work
in the following chapters focuses on defining the mechanism by which V. cholerae forms
positive feedback loops between QS regulators and the production of CAI-1.

10
A
B
C
D
E
Figure 1.1 Quorum-sensing autoinducers
A. N-3-(oxo-hexanoyl)-HSL (V. fischeri), 3-oxo-C12-HSL (P. aeruginosa), C4-HSL (P.
aeruginosa), and N-(3-hydroxybutyryl)-HSL (V. harveyi). B. 2-heptyl-3-hydroxy-4-
quinolone (P. aeruginosa). C. Autoinducing peptide I (S. aureus). D. 4,5-dihydroxy-2,3-
pentanedione (DPD) (universal autoinducer) and (2S,4S)-2-methyl-2,3,3,4-
tetrahydroxytetrahydrofuran borate (AI-2 in Vibrios). E. (Z)-3-aminoundec-2-en-4-one (V.
harveyi) and (S)-3-hydroxytridecan-4-one (V. cholerae).

11
A.
B.
Figure 1.2 Quorum sensing in V. fischeri
The V. fischeri LuxI-R QS system at A. LCD and B. HCD. “AHL” represents N-3-(oxo-
hexanoyl)-homoserine lactone and the double horizontal bars represent the inner and
outer bacterial membranes.

12
A.
B.
Figure 1.3 Quorum sensing in P. aeruginosa
The P. aeruginosa LasI-R/RhlI-R QS systems at A. HCD for the LasI-R system and B.
HCD for the RhlI-R system. The double horizontal bars represent the inner and outer
bacterial membranes.

13
A.
B.
Figure 1.4 Quorum sensing in S. aureus
The S. auerus QS system is represented at A. LCD and B. HCD. The horizontal bar
represents the bacterial membrane, and the “P” attached to AgrC and AgrA indicates
when these proteins are in the phosphorylated states. AgrD is a precursor for the
autoinducing peptide (AIP), which is processed and transported across the membrane
by AgrB. AgrC is a histidine kinase that acts as the receptor in a two component system
with the response regulator and transcription factor AgrA.

14
A.
B.
Figure 1.5 Quorum Sensing in V. harveyi
The QS system of V. harveyi at A. LCD and B. HCD. The double horizontal bars
represent the inner and outer bacterial membranes, “CAI-1” is (Z)-3-aminoundec-2-en-4-
one, AI-1 is N-(3-hydroxybutyryl)-homoserine lactone, and AI-2 is (2S,4S)-2-methyl-
2,3,3,4-tetrahydroxytetrahydrofuran borate, and the “P” attached to proteins indicates
when they are phosphorylated.

15
A.
B.
Figure 1.6 Quorum Sensing in V. cholerae
The QS system of V. cholerae at A. LCD and B. HCD. The double horizontal bars
represent the inner and outer bacterial membranes, the “P” attached to proteins
indicates when they are phosphorylated, “CAI-1” is (S)-3-hydroxytridecan-4-one, and AI-
2 is (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate.

16
CHAPTER 2:
REGULATION OF QS SYNTHASES AND RECEPTORS IN V. CHOLERAE

17
Introduction
Quorum sensing is a cell-to-cell communication process used by bacteria to
determine their population density and regulate community-dependent gene expression.
The human pathogen V. cholerae contains two parallel QS circuits that converge onto a
shared phosphorelay system. The autoinducer receptors, CqsS and LuxPQ, and their
cognate autoinducer synthases, CqsA and LuxS, control the V. cholerae QS response.
Although positive feedback from a master regulator to autoinducer production is a nearly
universal feature of bacterial QS circuits, regulation of the autoinducer synthase and
receptor genes in V. cholerae has not been explored. Here, we examined expression of
these the V. cholerae QS autoinducer synthase and receptor genes at the RNA and
protein levels in multiple QS-locked mutant backgrounds during the transition from
exponential to stationary phase. We demonstrate that positive feedback exists in the V.
cholerae QS system, but is confined only to the CqsA/CqsS QS circuit. The LuxS/LuxPQ
does not exhibit regulation under the conditions tested, suggesting a biased-feedback
circuit topology in the V. cholerae QS system.
Results
Positive feedback loops between a QS circuit master regulator and its partner
autoinducer production gene are a nearly ubiquitous feature of bacterial QS systems.
However, the existence of such a feature has not been investigated in V. cholerae. In
order to determine if the genes encoding the QS receptors or QS synthases of the V.
cholerae QS circuit are auto-regulated, we measured mRNA and protein levels from
these genes in QS mutant backgrounds locked in differing QS states. We also measured
gene expression over the course of growth, from exponential to stationary phase, to
determine if growth phase plays a role in QS gene expression.

18
LuxS and LuxQ are not regulated by QS or growth phase
Previous work has shown that the LuxS AI-2 synthase and the LuxPQ AI-2
receptor are present in diverse bacterial species. Nonetheless, despite being a crucial
component of the V. cholerae QS pathway, autoregulation of these genes has not yet
been investigated. To examine if feedback in the QS pathway regulates these genes,
luxS-FLAG and luxQ-FLAG C-terminal translational fusions were generated and the
genes cloned onto plasmids. These fusions are driven by their endogenous promoters,
enabling measurement of RNA via qPCR and protein via Western blotting. The plasmids
were introduced into wild-type and QS-locked strains of V. cholerae containing the ΔvpsL
mutation to minimize biofilm formation that could interfere with optical density
measurements. The low cell density (LCD) locked V. cholerae strain contains the
phosphomimetic luxO-D47E allele that is constitutively active and continuously activates
the promoters of qrr1-4. In this configuration, Qrr1-4 levels are high, and consequently
hapR RNA is degraded, so no HapR master regulator is produced. The high cell density
(HCD) locked strain contains the ΔluxO allele, and consequently the qrr1-4 promoters
are not activated. In this configuration, because qrr1-4 are not transcribed, HapR
production is constitutive. Because the growth phase of the bacteria can also affect
expression of genes, samples were collected at multiple points throughout the LCD to
HCD transition.
Cultures were back-diluted into shaking flasks and samples were processed for
RNA and protein assessment. We first observed that there is no difference in luxS
mRNA expression among the different strain backgrounds tested, suggesting that luxS
mRNA is not QS-regulated (Figure 2.1A). Additionally, luxS mRNA levels were
consistent among all growth phases tested, suggesting that there is also no growth-

19
phase regulation. LuxS protein levels were also consistent between wild-type and QS-
locked strain backgrounds (Figure 2.1B), further suggesting no QS regulation of LuxS.
Similarly, LuxS protein levels were identical regardless of growth phase.
In addition to the synthase LuxS, we also investigated QS and growth control of
LuxQ using mRNA and protein measurements in analogous experiments and strains.
Trends in luxQ RNA levels were identical to those for luxS (Figure 2.2A), indicating that
luxQ mRNA is not regulated by the V. cholerae QS circuit or by growth phase.
Additionally, LuxQ protein levels were also consistent between strain backgrounds and
between growth phases (Figure 2.2B).
CqsA and CqsS are QS and growth phase regulated
Autoregulation onto CqsA and CqsS from the QS circuit has not been
investigated. To test for regulation of the CqsA synthase and CqsS receptor, CqsA-
FLAG and CqsS-FLAG translational fusions were constructed on plasmids and
introduced into wild-type and QS-locked strains of V. cholerae. Like the LuxS-FLAG and
LuxQ-FLAG fusions mentioned earlier, the CqsA-FLAG and CqsS-FLAG fusions were
driven by their endogenous promoters.
Cultures were back-diluted into shaking flasks, and samples were isolated
throughout growth. cqsA mRNA expression increased during the transition from
exponential to stationary phase, suggesting that cqsA is growth-phase regulated (Figure
2.3A). Additionally, at the same ODs, cqsA mRNA levels differed between wild-type and
QS-locked strain backgrounds. Specifically, LCD locked strains exhibited lower than
wild-type levels of cqsA mRNA throughout the growth curve, whereas HCD locked
strains showed higher than wild-type levels of cqsA mRNA, also throughout the growth
curve. CqsA protein (Figure 2.3B) tracked with cqsA mRNA levels. Protein levels

20
increased in all cases from exponential to stationary phase. LCD and HCD locked strains
showed lower and higher production, respectively, compared to wild-type.
We also determined that cqsS mRNA and CqsS protein levels exhibited patterns
similar of those found for cqsA. cqsS mRNA increased in all strains during the transition
from exponential to stationary phase (Figure 2.4A). cqsS mRNA levels were higher in
HCD locked strains than in wild-type, and LCD locked strains possessed lower cqsS
mRNA than wild-type. Likewise, CqsS protein levels (Figure 2.4B) were identical to those
found for cqsS mRNA.
Discussion
The CqsA-CqsS and LuxS-LuxPQ QS systems in V. cholerae act in parallel to
regulate downstream genes responsible for individual and group behaviors (Miller et al.,
2002). Despite a clear understanding of the general topology of the circuit, specific
questions regarding feedback in these systems have not been investigated. Positive
feedback onto autoinducer production is a nearly universal feature of QS systems. This
feedback loop presumably exists to hasten the transition from the LCD to HCD state by
ensuring that bacterial populations are flooded with autoinducer after an initial increase
in autoinducer concentration occurs. Interestingly, we have shown that the architecture
of the V. cholerae QS system appears to contain biased-feedback in the parallel circuit.
We first demonstrated that the LuxS-LuxPQ QS system is neither QS nor growth
phase regulated. This feature of the QS circuit is striking because it is counter to the
ubiquitous feedback setup found in other QS systems. There are a number of potential
reasons why the V. cholerae LuxS-LuxPQ QS system does not contain feedback. First,
since LuxS is tied directly to SAM metabolism (Schauder et al., 2001), and thus, central
metabolism, positive feedback in this case may disrupt key concentrations of metabolites,

21
and thus needs to be avoided. A second possibility is that the LuxS-LuxPQ system lacks
positive feedback because the system cannot tolerate it. The LuxS-LuxPQ system
detects AI-2, a class of interconvertible molecules shown to be a generic bacterial QS
signal. In this case, V. cholerae may integrate bacteria-wide population density
information into it’s QS circuit, and thus amplification of AI-2 would skew its recognition
of other species in the local community.
We also demonstrated that the CqsA-CqsS system is both QS and growth phase
regulated. Unlike the LuxS-LuxPQ system, the CqsA-CqsS system follows the positive
feedback convention found in the architecture of most other QS circuits. We showed that
both mRNA and protein levels of these genes differ between locked LCD and HCD
strains, suggesting it is the mRNA that is regulated. The mRNA could be regulated at the
transcriptional or post-transcriptional level, and this will be investigated in future work.
Presumably, this feedback setup functions to rapidly push V. cholerae into a HCD state
after initial CAI-1 concentration thresholds are reached. In addition to QS feedback, both
cqsA and cqsS are regulated by growth phase. Neither gene is expressed in the wild-
type at exponential phase, but both are fully expressed at stationary phase. One
explanation for this may be that V. cholerae promotes individual behaviors if nutrient
conditions are such that rapid growth is possible, even in instances when the population
size is large.
The exact mechanism for CAI-1 feedback has not been defined at this point and
merits further investigation. The feedback could potentially operate similarly to LuxI-LuxR
systems, in which HapR could activate cqsA in order to produce more CAI-1.
Alternatively, the feedback could resemble the LuxMN positive feedback system in V.
harveyi, in which Qrr sRNAs negatively regulate luxMN expression at LCD, creating a
positive feedback loop as Qrr levels decrease when entry into HCD occurs. Future work

22
will focus on further delineating the specific mechanism behind feedback in the CqsA-
CqsS QS system.

23
A. B. Figure 2.1 LuxS is not regulated by growth-phase or QS
(A) luxS mRNA was measured by qRT-PCR and (B) LuxS protein was measured by
Western blotting in V. cholerae from a luxS-FLAG translational fusion in the ΔvpsL,
ΔvpsL luxO-D47E (locked LCD), and the ΔvpsL ΔluxO (locked HCD) strains. Samples
were isolated from the same flask at different points during growth.

24
A. B. Figure 2.2 LuxQ is not regulated by growth-phase or QS (A) luxQ mRNA was measured by qRT-PCR and (B) LuxQ protein was measured by
Western blotting in V. cholerae from a luxQ-FLAG translational fusion in the ΔvpsL,
ΔvpsL luxO-D47E (locked LCD), and ΔvpsL ΔluxO (locked HCD) strains. Samples were
isolated from the same flask at different points during growth.

25
A. B. Figure 2.3 CqsA is growth-phase and QS regulated (A) cqsA mRNA was measured by qRT-PCR and (B) CqsA protein was measured by
Western blotting in V. cholerae from a cqsA-FLAG translational fusion in the ΔvpsL,
ΔvpsL luxO-D47E (locked LCD), and ΔvpsL ΔluxO (locked HCD) strains. Samples were
isolated from the same flask at different points during growth.

26
A. B. Figure 2.4 CqsS is growth-phase and QS regulated
(A) cqsS mRNA was measured by qRT-PCR and (B) CqsS protein was measured by
Western blotting in V. cholerae from a cqsS-FLAG translational fusion in the ΔvpsL,
ΔvpsL luxO-D47E (locked LCD), and ΔvpsL ΔluxO (locked HCD) strains. Samples were
isolated from the same flask at different points during growth.

27
Materials and Methods
Bacterial strains and media
V. cholerae El Tor C6706str2 and isogenic mutant strains and E. coli S17-1λpir
(de Lorenzo and Timmis, 1994) strains were grown at 30°C in Luria-Bertani (LB) medium.
Liquid cultures were grown in flasks or test tubes with shaking for aeration. Strains used
in this study are noted in Table 2.1. Antibiotics were used at the following concentrations:
chloramphenicol 10 µg/mL, polymyxin B 50 U/mL, and streptomycin 5mg/mL. Plasmids
were electroporated into electrocompetent E. coli S17-1λpir by the MicroPulser (Bio-
Rad).
DNA manipulations and plasmids
Plasmid construction was performed by standard methods. Polymerase chain
reactions (PCR) were performed using the iProof DNA polymerase (Bio-Rad), and
restriction digestions and ligations were performed using restriction endonucleases and
T4 DNA ligase (New England Biolabs).
Plasmids pZND86, pZND87, pZND88, and pZND95 were constructed by a
similar strategy. All plasmids contain synthase or receptor promoters and ORFs PCR-
amplified from the V. cholerae El Tor C6706str2 genome. The reverse primer in each
case encodes the C-terminal FLAG fusion, and the T1 stem loop of the E. coli rrnB rho-
independent terminator (Table 2.2). Specifically, plasmid pZND86 contains 507 base
pairs upstream of the cqsS translational start site and the cqsS ORF. Plasmid pZND87
contains 500 base pairs upstream of the luxS translational start site and the luxS ORF.
Plasmid pZND88 contains 488 base pairs upstream of the luxP translational start site
and the luxP ORF and luxQ ORF. Plasmid pZND95 contains 522 base pairs upstream of

28
the cqsA translational start site and the cqsA ORF. PCR amplicons for pZND87, pZND88,
and pZND95 were digested with SalI and BamHI restriction endonucleases and ligated
into pJS1194 digested with the same endonucleases. The pZND86 PCR amplicon was
digested with NotI and KpnI restriction endonucleases and ligated into pJS1194 digested
with the same endonucleases. Plasmids were transformed into E. coli S17-1λpir and
mated to V. cholerae strains as described (Skorupski and Taylor, 1996).
RNA and protein isolation
Overnight cultures were back-diluted 1:1000 into fresh LB and shaken in flasks.
OD measurements were conducted periodically, and RNA and protein samples were
isolated simultaneously at OD = 0.2, 0.6, 1.0, and 1.4. For RNA samples, 5 OD units of
culture were added to 20% RNA stop solution (95% ethanol, 5% phenol), mixed by
inversion, and frozen in liquid nitrogen. Samples were stored at -80°C until processing
the following day. For protein samples, 1 OD unit of culture was immediately centrifuged
at 16,000xg for 1 minute, the supernatant was removed, and pellets were stored at
-80°C until further processing.
To isolate RNA from the samples, samples were thawed at room temperature
and centrifuged at 5000x g for 10 minutes at 4°C. Supernatants were discarded and
pellets were processed to extract RNA following the Trizol method (Tu and Bassler,
2007). To prepare protein from pellets, pellets were dissolved in Bug Buster (Millipore)
and incubated at room temperature for 20 minutes.

29
qRT-PCR
Following total RNA isolation, cDNA was generated from 1µg RNA using
Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s guidelines.
Quantitative real time PCR (qRT-PCR) analyses were conducted in quadruplicate from
each cDNA sample using Sybr green PCR master mix (Applied Biosystems) according
to Tu & Bassler, 2007. Measurements were performed on the ABI Prism Sequence
Detection System using the ΔΔCt method. Samples were normalized to an internal rpsL
control. All primers in this analysis can be found in Table 2.2.
Western blots
Samples prepared in BugBuster solution were mixed with 0.5% SDS. In
experiments testing for CqsA-FLAG or LuxS-FLAG protein levels, the BugBuster+SDS
mix was boiled for 3 minutes before being run on an SDS-PAGE gel. In experiments
testing for CqsS-FLAG or LuxQ-FLAG protein levels, 1% Triton-X was added to help
dissolve membrane-bound dimers, and the BugBuster+SDS mix was incubated at 37°C
for 1 hour before being applied to a SDS-PAGE gel. 10µl of each sample was
electrophoresed at 150V for 2 hours on 15-well 4-15% gradient SDS-PAGE gels (Bio-
Rad). Experiments testing for CqsA-FLAG were wet blotted onto nitrocellulose
membranes, and experiments testing for CqsS-FLAG, LuxS-FLAG, and LuxQ-FLAG
were wet blotted onto PVDF membranes pre-soaked in methanol. Gels were blotted for
1 hour at 100V at 4°C. After blotting, membranes were cut in half to allow the RNA
polymerase β’ subunit control and FLAG epitopes to be probed independently. After
blotting, membranes were blocked in TBST+5% dry milk for 1 hour with shaking at room
temperature. Subsequently, α-FLAG-HRP antibody (Sigma-Aldrich) was diluted 1:5000

30
in TBST and incubated against the FLAG membrane for 2 hours. Simultaneously, α-RNA
polymerase β’ subunit antibody (Abcam) was diluted 1:100,000 in TBST and incubated
against the RNA polymerase β’ subunit membrane for 1 hour. The membranes testing
for α-RNA polymerase β’ subunit were rinsed with TBST and incubated with α-mouse-
HRP for 1 hour. After this incubation, both membranes containing the FLAG and α-RNA
polymerase β’ subunit were rinsed with TBST and incubated with Amersham EL Prime
Western Blotting Detection Reagent according to the manufacturer’s guidelines.
Membranes treated with the chemiluminescent reagents were visualized via film
exposure for 5 minutes.

31
Plasmid name Description Source pJS1194 Empty vector J. Schaffer, unpublished pZND86 pJS1194 with cqsS-FLAG This study pZND87 pJS1194 with luxS-FLAG This study pZND88 pJS1194 with luxPQ-FLAG This study pZND95 pJS1194 with cqsA-FLAG This study
Table 2.1 Plasmids used in this study

32
Table 2.2 Primers used in this study
Primer name Sequence Use 401 AAA AGC GGC CGC CGC TAT TTA CTC AAA CGT AAA GAG G pZND86 (insert) 404 AAA AGC GGC CGC GCA TGC AAA AAG ACC CTT CAT AAA T pZND86 (vector) 405 AAA AGT CGA CGG CGT AAA GTG GTA CTG GAA GGT C pZND87 (insert)
406 AAA AGG ATC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTT GTC GTC ATC GTC TTT GTA GTC GTG AAC CTT CAG CTC ATT GAG CA
pZND87 (insert)
407 AAA AGT CGA CCT TTA TCG CCG CGG TGG ATC TTG pZND88 (insert)
408 AAA AGG ATC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTT GTC GTC ATC GTC TTT GTA GTC ATT TAA GCC AGC GTT TTT TTG GCC
pZND88 (insert)
412 AAA AGG ATC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTT GTC GTC ATC GTC TTT GTA GTC ACG AAA ATA AAA ATC ACC GTA GTT GAC CG
pZND95 (insert)
419 AAA AGG TAC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTT GTC GTC ATC GTC TTT GTA GTC CAC CCA AGC TGC CAC TTT ATT TAG C
pZND86 (insert)
420 AAA AGG TAC CGG ATC CGG TGA TTG ATT GAG CAA GC pZND86 (vector) 422 CGT CAT CGT CTT TGT AGT C FLAG qRT-PCR 424 TCG CTT ATC ACT CAA TAG TG cqsA qRT-PCR 471 CAA AAC CTT GGC TCT GGT AC cqsS qRT-PCR 472 CGG AAT ATG TGT TAG TGA AGC luxQ qRT-PCR 473 GAT TGC GAA AAA CGT GAT TGC luxS qRT-PCR
MB51 AAA AGT CGA CTT GCG CAG CCC GAC CCG ATT C pZND95 (insert)
VC597 GGT GTT CGC TAC CAC ACA GTT rpsL forward qRT-PCR
VC598 AAG ACT TAG GAC GCT TCA CAC C rpsL reverse qRT-PCR

33
Strain name Organism Genotype Plasmid Source
CW2034 V. cholerae C6706str2 ΔvpsL None (Waters et al., 2008)
CW2035 V. cholerae C6706str2 ΔvpsL luxO-D47E None (Waters et al., 2008)
CW2037 V. cholerae C6706str2 ΔvpsL ΔluxO None (Waters et al., 2008)
ZDC424 V. cholerae C6706str2 ΔvpsL pZND87 This study ZDC426 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND87 This study ZDC430 V. cholerae C6706str2 ΔvpsL ΔluxO pZND87 This study ZDC432 V. cholerae C6706str2 ΔvpsL pZND88 This study ZDC434 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND88 This study ZDC438 V. cholerae C6706str2 ΔvpsL ΔluxO pZND88 This study ZDC450 V. cholerae C6706str2 ΔvpsL pZND86 This study ZDC454 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND86 This study ZDC462 V. cholerae C6706str2 ΔvpsL ΔluxO pZND86 This study ZDC473 V. cholerae C6706str2 ΔvpsL pZND95 This study ZDC475 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND95 This study ZDC479 V. cholerae C6706str2 ΔvpsL ΔluxO pZND95 This study
Table 2.3 Bacterial strains used in this study

34
CHAPTER 3:
IDENTIFICATION OF REGULATORS OF CQSA AND CQSS IN V. CHOLERAE

35
Introduction
Quorum sensing is a mechanism of cell-to-cell communication used by bacteria
to determine their population size and regulate the expression of genes involved with
group behaviors. In the Vibrio cholerae QS system, the autoinducer synthases CqsA and
LuxS catalyze the production of autoinducers CAI-1 and AI-2 which bind the autoinducer
receptors CqsS and LuxPQ. These parallel QS circuits integrate through a shared two-
component phosphorelay system. The CqsA/CqsS QS system is regulated in a manner
consistent with a positive feedback loop originating from a QS regulator. Since multiple
regulators exist in the V. cholerae QS system, we used genetic methods to identify the
regulator responsible for positive feedback onto the CqsA/CqsS system. We show that
the Qrr1-4 sRNAs are responsible for regulating CqsA, and that this regulation occurs
indirectly through an unidentified intermediate.
Results
cqsA and cqsS regulated by the Qrr sRNAs
The preceding chapter showed that cqsA and cqsS mRNA and CqsA and CqsS
protein production are different between wild-type V. cholerae and LCD and HCD-locked
mutant strains. We aimed to define the components involved in this regulation. First, we
considered known QS regulators as candidates, and we conducted epistasis
experiments. A tetracycline inducible qrr4 gene on a plasmid was introduced into four V.
cholerae mutants harboring combinations of single, double, and triple deletions of the
three known QS regulators. cqsA and cqsS mRNA were measured in the ΔluxO single
mutant, the ΔluxO ΔhapR and ΔluxO ΔaphA double mutants, and the ΔluxO ΔaphA
ΔhapR triple mutant with and without induction of qrr4 expression. cqsA expression

36
decreased following induction of Qrr4 in all four strains, suggesting that cqsA regulation
is independent of AphA and HapR, and thus is Qrr4-dependent (Figure 3.1). cqsS
expression modestly decreased in all strains following Qrr4 induction, however, the fold
change was not as substantial as that for cqsA (Figure 3.2).
To further verify that cqsA and cqsS are Qrr-regulated, we measured cqsA and
cqsS mRNA levels in additional QS-mutant strains. (Jian-Ping Cong, unpublished data).
cqsA and cqsS mRNA levels were measured in luxO-D47E vs. luxO-D47E Δqrr1-4
strains to verify that regulation of cqsA and cqsS is a result of Qrr regulation and not
direct LuxO regulation. cqsA and cqsS expression was higher in the luxO-D47E Δqrr1-4
strain than in the luxO-D47E strain. Thus, the Qrr sRNAs are required for cqsA and cqsS
regulation. All strains lacking HapR showed no change, verifying that cqsA and cqsS are
not regulated by HapR (not shown). Going forward, we are using cqsA as a proxy for
cqsS because cqsA regulation by Qrr4 is more dramatic than that for cqsS.
Qrr4 indirectly regulates cqsA and cqsS
The Qrr sRNAs function by binding to complementary target mRNA sequences,
generally near translational start sites and catalyzing degradation or sequestration of the
mRNAs. Because Qrr4 repressed the expression of cqsA and cqsS mRNA, we
attempted to identify sequences in or near these genes complementary to Qrr4. We first
used 5’ RACE and RNASeq to define the start and end of the cqsA and cqsS transcripts
to determine what sequences could be available for base pairing. We found that the
5’UTR of cqsA is 41bp long, and the 5’UTR of cqsS is 294bp long. We used the
bioinformatic tool RNAHybrid and visual inspection to scan for complementary
sequences between cqsA/S and Qrr4, but no complementary sequences were present.

37
To test for direct Qrr4 regulation of the cqsA transcript, we constructed a two-
plasmid system in E. coli, which does not contain V. cholerae QS components. One
plasmid carried tetracycline-inducible qrr4 and the second plasmid harbored an IPTG-
inducible cqsA-mKate2 translational fusion. Our rationale was that if regulation was
direct, Qrr4 would be sufficient to control expression of cqsA in E. coli. cqsA expression
did not change with increasing Qrr4 induction, indicating that cqsA is not directly
regulated by Qrr4 (Figure 3.3).
To further verify that Qrr regulation of cqsA is indirect, we conducted a Qrr4-pulse
experiment in vivo in V. cholerae to measure the timing for Qrr4 to repress cqsA
expression. Our rationale was that posttranscriptional regulation occurs rapidly, within
two minutes. Thus, if cqsA is regulated indirectly, the timescale for regulation would be
longer than what is typical for direct regulation. We pulse-induced Qrr4 in the ΔluxO
strain and measured cqsA mRNA by Northern blotting. 2-fold repression of cqsA
occurred only after 30 minutes of Qrr4 induction, suggesting that Qrr4 likely acts through
an intermediate to regulate cqsA (Figure 3.4).
A candidate screen for cqsA regulators
We reasoned that if Qrr4 acts indirectly to control cqsA, it must act through an
intermediate. Previously, a screen was conducted in V. cholerae to identify genes
regulated by Qrr4. Specifically, Qrr4 was pulse-induced, and V. cholerae gene
expression was measured by microarray analysis (Yi Shao, Table 3.1). 18 genes
identified in this microarray are candidates for regulatory intermediates acting between
Qrr4 and cqsA. If so, our expectation was that a deletion in an intermediate acting
between Qrr4 and cqsA would be incapable of transmitting information about Qrr
concentration to cqsA/S. We hypothesized that such a mutant would show a cqsA HCD

38
phenotype because the mutant could not repress cqsA. To test for cqsA regulation by
these 18 candidates, a plasmid containing cqsA-FLAG driven by its endogenous
promoter was introduced into V. cholerae strains each containing a deletion in one of the
candidate genes regulated by Qrr4. CqsA-FLAG expression in these strains was
compared to that in V. cholerae wild-type, locked LCD, and locked HCD strains, and six
strains exhibited cqsA expression similar to the locked HCD strain (Figure 3.5A). Three
of these strains contained deletions in hypothetical genes (VCA0935, VC1280, and
VC1323), two contained deletions in ABC transporter genes (VC1327 and VC0704), and
one contained a deletion in a galactokinase (VC1595). Because their phenotypes were
similar to wild-type, the other 12 strains were not further examined. We next introduced a
plasmid containing tetracycline-inducible qrr4 into the six HCD phenotype strains to
attempt to complement the apparent cqsA expression defects (Figure 3.5B). Our
rationale was that if the strain with a deletion in the putative intermediate remained
capable of regulation of cqsA via qrr4 induction, then the candidate gene could not, in
fact, be the correct intermediate. All strains showed decreased cqsA expression
following qrr4 induction, suggesting that Qrr4 was still capable of regulating cqsA. Thus,
none of the genes from the microarray appear to be the intermediate factor that links
Qrr4 to cqsA.
Discussion
The genes encoding the V. cholerae autoinducer synthase and receptor, cqsA
and cqsS, are activated during the transition from LCD to HCD. The preceding chapter
has demonstrated that this change in expression is controlled by QS. However, the
specific mechanism was not defined. We examined four QS genes (hapR, aphA, luxO,
and qrr4) for potential roles in cqsA and cqsS regulation. There are multiple models that

39
could explain how cqsA and cqsS are QS regulated. For example, cqsA/S could be
activated during the transition from LCD to HCD by HapR, which increases in
concentration from LCD to HCD. Alternatively, cqsA/S could be repressed by AphA or
Qrr1-4 at LCD, and the amount of repression would decrease during the transition from
LCD to HCD as these regulators decrease in concentration. We devised genetic
epistasis experiments to determine which components of the QS circuit influence cqsA/S
regulation. These experiments showed that the QS-based regulation of cqsA/S
originates from the Qrr sRNAs, and regulation can be controlled by exogenous
production of Qrr4. Even in mutants in which the two QS master regulators (HapR and
AphA) were deleted, cqsA/S expression was modified by addition of Qrr4.
Qrr1 and Qrr5 were previously shown to repress luxMN expression in the related
species V. harveyi by directly binding and catalyzing degradation of the mRNA (Teng et
al., 2011). We were surprised to find that although cqsA/S (a parallel synthase/receptor
pair) are also Qrr-regulated, the regulation in this case is not direct. No apparent Qrr-
binding sites could be found in either cqsA or cqsS, and we showed that a cqsA-mKate2
translational fusion is not regulated by Qrr4 in E. coli. We also showed that cqsA
repression by Qrr4 pulse-induction in V. cholerae occurs only after 32 minutes, even
though direct regulation should occur in only a few minutes. Although the V. harveyi and
V. cholerae QS circuits are highly similar, many differences have arisen and this finding
is yet another. A similar situation exists with respect to LuxR/HapR positive feedback
onto the Qrr sRNAs. LuxR directly binds the qrr1-5 promoters and activates expression
in V. harveyi, whereas HapR indirectly activates qrr1-4 though an unknown intermediate
in V. cholerae (Svenningsen et al., 2009). These results indicate that V. cholerae uses
more steps between autoregulatory loops in the QS circuit than does V. harveyi.
Possibly, such a regulatory architecture is critical for proper timing of regulation, or it

40
could provide additional points through which additional information from other regulatory
pathways can be integrated.
We devised a model that accounts for the indirect Qrr regulation of cqsA. We
propose two possibilities (Figure 3.6). In one scheme, Qrr1-4 activate a regulator ‘Y’ that
represses cqsA. Alternatively, Qrr1-4 represses a regulator ‘X’ that activates cqsA. In
both cases, induction of Qrr sRNAs ultimately results in repression of cqsA. Our
analyses of candidates for X and Y showed that deletions in six Qrr4-regulated genes
displayed locked cqsA HCD phenotypes similar to that of the ΔluxO strain, but in these
candidate mutants cqsA remained regulated by exogenous production of Qrr4. It is
possible that these genes may be involved in altering endogenous Qrr levels. It is also
possible that there are multiple steps between the Qrr sRNAs and cqsA, and therefore
this screen only accounts for the first step in this chain of regulation. Finally, it is also
possible that the true intermediate that acts between Qrr4 and cqsA did not change
expression strongly enough to be identified through the microarray screen, and therefore
was not included in the list of 18 candidates. In order to better understand the
mechanism that underlies Qrr4 regulation of cqsA, the future studies will examine how
cqsA is regulated using cqsA fusion constructs.

41
A.
B.
Figure 3.1 cqsA and cqsS are regulated by Qrr4
A. cqsA and B. cqsS mRNAs were measured by qRT-PCR in V. cholerae QS deletion
mutants. Blue bars show no qrr4 induction. 25ng/mL aTc was added to induce qrr4
expression from a tetracycline-inducible promoter on a plasmid (yellow bars). Samples
were isolated at OD600 = 1.0. Four replicates were measured in each sample; bar height
represents the mean and error bars represent standard error of the mean (SEM).

42
A.
B.
Figure 3.2 cqsA and cqsS are regulated by the Qrr sRNAs
A. cqsA and B. cqsS mRNAs were measured using the Quantgene plex reagent system
in V. cholerae QS mutants. Three independent measurements were taken and mean
and SEM are shown.

43
Figure 3.3 Qrr4 does not directly regulate cqsA
CqsA-mKate2 production was measured by FACS. Anhydrous tetracycline (aTc) was
added to LB at concentrations of 0 ng/mL, 10 ng/mL, 50 ng/mL, and 100ng/mL to induce
qrr4 expression. IPTG was added at concentrations of 0uM, 10uM, 100uM and 500uM to
induce cqsA-mKate2 expression.

44
Figure 3.4 Qrr4 pulse-induction regulation of cqsA
cqsA and Qrr4 mRNAs were measured by Northern blotting. A culture of V. cholerae
harboring Ptet-qrr4 was divided into aliquots at OD600 = 1.0, and 50ng/mL aTc was
added to one aliquot to induce qrr4 expression. Samples from both aliquots were
removed after 0, 2, 4, 8, 16, and 32 minutes, and RNA was isolated.

45
A.
B.
Figure 3.5 Screen for cqsA regulators
A. cqsA-FLAG expression was measured by Western Blotting in strains containing
deletions of 18 genes regulated by Qrr4. All samples were isolated at OD600 =1.0. B.
cqsA-FLAG expression was measured by Western Blotting in strains exhibiting locked
HCD cqsA phenotypes. “-“ indicates no Qrr4 induction by aTc and “+” indicates Qrr4
induction by 50ng/mL aTc following back-dilution from overnight cultures.

46
Figure 3.6 Model for Qrr regulation of cqsA in V. cholerae
The QS circuit of V. cholerae is shown. At LCD, CqsS acts as a kinase, and the
downstream phosphorelay activates the expression of the Qrr sRNAs. At HCD, CqsS
acts as a phosphatase, and the Qrr sRNAs are not activated. Qrr1-4 could feedback
onto CqsA via an intermediate “X” or “Y.”

47
Materials and Methods
Bacterial strains and media
V. cholerae El Tor C6706str2 and isogenic mutant strains were grown at 30°C
and E. coli S17-1λpir (de Lorenzo and Timmis, 1994) strains were grown at 37°C in
Luria-Bertani (LB) medium. Liquid cultures were grown in flasks or test tubes with
shaking for aeration. Strains used in this study are noted in Table 3.3. Antibiotics were
used at the following concentrations: chloramphenicol 10 µg/mL, ampicillin 100ug/mL.
Plasmids were electroporated into electrocompetent E. coli S17-1λpir by the MicroPulser
(Bio-Rad).
DNA manipulations and plasmids
Plasmid construction was performed by standard methods. Polymerase chain
reactions (PCR) were performed using the iProof DNA polymerase (Bio-Rad), and
restriction digestions and ligations were performed using restriction endonucleases and
T4 DNA ligase (New England Biolabs). Plasmid pZND97 was constructed by PCR-
amplifying cqsA 41bp upstream of the ORF from V. cholerae C6706str2 gDNA and the
mKate2 ORF from pZND79. The two amplicons were combined via splicing by overlap
extension (SOEing) PCR. The vector for pZND97 was created by PCR-amplifying the
backbone of pZE12. The amplicons were digested with BamHI and semi-blunt ligated
together. Plasmid pZND104 was created by amplifying the Ptet-qrr4 region on pZA-qrr4
and subcloning onto the pASK75 vector. Specifically, 54bp upstream of the qrr4
transcriptional start site though the qrr4 rho-independent terminator were amplified. The
amplicon and pASK75 were digested with AvrII and KpnI-HF and ligated together.
Plasmid pZND122 was created by amplifying the 493bp transfer origin oriT of pEVS143

48
and subcloning this region onto pZND104. Specifically, the oriT region was PCR-
amplified from pEVS143 and the vector backbone was amplified from pZND104, and the
amplicons were digested with KpnI-HF and XbaI-HF and ligated together. The aphA
deletion plasmid pZND46 was constructed by PCR-amplifying regions directly upstream
(769bp) and downstream (800bp) of the aphA ORF and fusing these amplicons together
by SOEing PCR. This region was subcloned onto pKAS32 by digesting the insert and
vector with NotI and EcoRI restriction endonucleases (NEB) and ligating together. All
plasmids were transformed into E. coli S17-1λpir, and V. cholerae-harbored plasmids
were mated to V. cholerae strains as described (Skorupski and Taylor, 1996).
qRT-PCR
Overnight cultures were back-diluted 1:1000 into fresh LB and shaken in flasks at
30°C, and OD measurements were conducted periodically. Cell cultures were isolated at
OD=1.0, and 5 OD units of culture were added to 20% RNA stop solution (95% ethanol,
5% phenol), mixed by inversion, and frozen in liquid nitrogen. Samples were stored at -
80°C until processing the following day. To isolate RNA from the samples, samples were
thawed at room temperature and centrifuged at 5000x g for 10 minutes at 4°C.
Supernatants were discarded and pellets were processed to extract RNA following the
Trizol method (Tu and Bassler, 2007).
Following total RNA isolation, cDNA was generated from 1µg RNA using
Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s guidelines.
Quantitative real time PCR (qRT-PCR) analyses were conducted in quadruplicate from
each cDNA sample using Sybr green PCR master mix (Applied Biosystems) according
to Tu & Bassler, 2007. Measurements were performed on the ABI Prism Sequence

49
Detection System using the ΔΔCt method. Samples were normalized to an internal rpsL
control. All primers in this analysis can be found in Table 3.2.
Northern Blots
cqsA riboprobes were synthesized by first PCR-amplifying V. cholerae El Tor
C6706str2 gDNA template with primers in Table 3.2, and then performing T7 in vitro
transcription on the PCR amplicon (Ambion) with 32P-α-UTP. qrr4 oligoprobes were
synthesized by end-labeling primers in Table 3.2 with 32P-γ-ATP using T4 PNK (New
England Biolabs). Riboprobes and oligoprobes were purified by Illustra Microspin
columns according to the manufacturer’s guidelines (GE Healthcare). For Northern blot
experiments, an overnight culture was back-diluted 1:1000 into fresh LB and shaken in a
flask at 30°C, was grown to OD=1.0, and subsequently split into two flasks each
containing half of the volume of the original. 50ng/mL aTc was added to one flask, and
50ul of ethanol was added to the other. 5 OD units of culture were isolated from each
flask at 2, 4, 8, 16, and 32 minutes after reaching OD=1.0. RNA was Trizol extracted as
above. 10ug of total RNA was resolved on 6% polyacrylamide (PAA, 7M urea) at 300V
for 2 hours. RNA was transferred to Amersham Hybond-XL nylon membranes
(Amersham Biosciences, Piscataway, NJ) for 1 hour at 50V and at 4°C. For hybridization,
membranes were incubated overnight at 70°C for cqsA riboprobes and at 42°C for qrr4
oligoprobes in 15mL Rapid-hyb buffer (GE Healthcare, Piscataway, NJ). Following
overnight incubation, membranes were washed three times in SSC buffer + 0.1%SDS
(5X, 1X, and 0.5X at 42°C for oligoprobes; 2X, 1X, and 0.5X at 70°C for riboprobes).
Blots were imaged for 20 hours unless otherwise noted on phophorimager screens and
scanned on a Typhoon 9410 (GE Healthcare).

50
Direct RNA measurements
RNA amounts were measured using the Quantigene plex 2.0 reagent system
(Affymetrix, Santa Clara, CA) as described (Tu et al., 2010). Overnight cultures were
back-diluted 1:1000 into fresh LB and shaken in flasks at 30°C, and OD measurements
were conducted periodically. Cell cultures were isolated at OD=1.0, and RNA was
processed following the manufacturer’s guidelines. Fluorescent beads targeting cqsA,
cqsS were normalized to beads targeting hfq.
Western blots
Overnight cultures were back-diluted 1:1000 into fresh LB and shaken in flasks at
30°C, and OD measurements were conducted periodically. Cell cultures were isolated at
OD=1.0, and 1 OD unit of culture was immediately centrifuged at 16,000xg for 1 minute,
the supernatant was removed, and pellets were stored at -80°C until further processing.
To prepare protein from pellets, pellets were dissolved in Bug Buster (Millipore) and
incubated at room temperature for 20 minutes. Samples prepared in Bug Buster solution
were mixed with 0.5% SDS. The Bug Buster+SDS mix was boiled for 3 minutes before
being run on an SDS-PAGE gel. 10µl of each sample was electrophoresed at 150V for 2
hours on 15-well 4-15% gradient SDS-PAGE gels (Bio-Rad). Protein was wet blotted
onto nitrocellulose membranes for 1 hour at 100V at 4°C. After blotting, membranes
were cut in half to allow the RNA polymerase β’ subunit control and FLAG epitopes to be
probed independently. After blotting, membranes were blocked in TBST+5% dry milk for
1 hour with shaking at room temperature. Subsequently, α-FLAG-HRP antibody (Sigma-
Aldrich) was diluted 1:5000 in TBST and incubated against the FLAG membrane for 2
hours. Simultaneously, α-RNA polymerase β’ subunit antibody (Abcam) was diluted

51
1:100,000 in TBST and incubated against the RNA polymerase β’ subunit membrane for
1 hour. The membranes testing for α-RNA polymerase β’ subunit were rinsed with TBST
and incubated with α-mouse-HRP for 1 hour. After this incubation, both membranes
containing the FLAG and α-RNA polymerase β’ subunit were rinsed with TBST and
incubated with Amersham EL Prime Western Blotting Detection Reagent according to
the manufacturer’s guidelines. Membranes treated with the chemiluminescent reagents
were visualized via film exposure for 5 minutes.
mKate2 reporter assay
E. coli strains were grown aerobically overnight at 37°C in LB medium. Cultures
were back-diluted 1:1000 into M9 + 0.5% glucose and grown to OD=0.1. Upon back-
dilution, anhydrous tetracycline (aTc, Clontech) or isopropyl β-D-1-thiogalactopyranoside
(IPTG) was added to cultures at concentrations indicated in Figure 3.3. mKate2
fluorescence was measured using flow cytometry (BD Biosciences FACSAria).

52
Table 3.1 Plasmids used in this study
Plasmid name Description Source pASK75 Empty vector (Skerra, 1994) pEVS143 Empty vector (Dunn et al., 2006)
pKAS32 Empty vector (Skorupski and Taylor, 1996)
pZA-qrr4 Ptet-qrr4 L. Feng, unpublished pZE12 Empty vector (Levine et al., 2007) pZND104 pASK75 with Ptet-qrr4 This study pZND122 pASK75 with Ptet-qrr4 and oriT This study pZND86 pJS1194 with cqsS-FLAG This study pZND95 pJS1194 with cqsA-FLAG This study pZND97 pZE12 with Ptac-cqsA-mKate2 This study pZND46 pKAS32-ΔaphA This study

53
Primer name Sequence Use 263 AAA AGC GGC CGC TGC GGG TGA AGC GAT CCA AAT T pZND46 (insert)
264 GTT TGG CTT GGC CCT CTA TCT AAA CGG TTG TTG TGC T pZND46 (insert) 265 GTT TAG ATA GAG GGC CAA GCC AAA CCT GTC GAT GTA pZND46 (insert) 266 AAA AGA ATT CCT GGC CAA CCG TTT GAA CAC TG pZND46 (insert) 411 /5Phos/GTC AGC TGG CGT TAA ATT TTT TAT AAC TAG pZND97 (insert) 429 AAA AGG ATC CTG CGG CGA GCG GTA TCA GCT CA pZND97 (vector) 430 GTG CTC AGT ATC TTG TTA TCC GC pZND97 (vector)
433 AAA AGG ATC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA TCT GTG CCC CAG TTT GCT AGG G pZND97 (insert)
485 GGC CTG AGA CCA GAA TTC GAG CT pZND104 (vector)
486 AAA AAC CTA GGA GGA ATT AAT CAT CTG GCC ATT CGA TGG pZND104 (vector)
497 AAA AAC CTA GGT CCC TAT CAG TGA TAG AGA TTG ACA TCC CTA T pZND104 (insert)
498 AAA AAG GTA CCA AGA TGC TAT GGC GAA TGT GGT GAA TA pZND104 (insert)
515 AAA AAA GGT ACC GAT CCG GTG ATT GAT TGA GCA AGC TTT pZND122 (insert)
516 AAA AAA TCT AGA AGC ACC GCC AGG TGC GAA TAA G pZND122 (insert)
517 AAA AAA TCT AGA CTG GAA CAA CAC TCA ACC CTA TCT CGG pZND122 (vector)
518 GCT TAT TAA CCA CCG AAC TGC GGG T pZND122 (vector)
539 CCA GCC CAA TAC GAA TGT TT cqsA riboprobe amplicon
540 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG CAA TGA TCC CAG GAC CAT GAC G
cqsA riboprobe amplicon
KPO-0063 CGT CTA TAA GTG TGA ACA ATG GTG qrr4 oligoprobe
Table 3.2 Primers used in this study

54
Strain name Organism Genotype Plasmid Source
ZDE302 E. coli S17-1λpir Wild type pZND97 This study ZDE339 E. coli BW-RI Wild type pZND97 This study ZDE347 E. coli BW-RI Wild type pZND97, pZA-qrr4 This study ZDC550 V. cholerae C6706str2 ΔvpsL ΔluxO pZND95, pZND122 This study ZDC691 V. cholerae C6706str2 ΔvpsL ΔluxO ΔhapR pZND95, pZND122 This study ZDC697 V. cholerae C6706str2 ΔvpsL ΔluxO ΔaphA pZND95, pZND122 This study
ZDC693 V. cholerae C6706str2 ΔvpsL ΔluxO ΔhapR ΔaphA pZND95, pZND122 This study
ZDC546 V. cholerae C6706str2 ΔvpsL ΔluxO pZND86, pZND122 This study ZDC687 V. cholerae C6706str2 ΔvpsL ΔluxO ΔhapR pZND86, pZND122 This study ZDC695 V. cholerae C6706str2 ΔvpsL ΔluxO ΔaphA pZND86, pZND122 This study
ZDC689 V. cholerae C6706str2 ΔvpsL ΔluxO ΔhapR ΔaphA pZND86, pZND122 This study
ZDC473 V. cholerae C6706str2 ΔvpsL pZND95 This study ZDC475 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND95 This study ZDC479 V. cholerae C6706str2 ΔvpsL ΔluxO pZND95 This study ZDC581 V. cholerae C6706str2 ΔVC0583 pZND95 This study ZDC582 V. cholerae C6706str2 ΔVC0704 pZND95 This study ZDC583 V. cholerae C6706str2 ΔVC0811 pZND95 This study ZDC584 V. cholerae C6706str2 ΔVC0975 pZND95 This study ZDC585 V. cholerae C6706str2 ΔVC0976 pZND95 This study ZDC586 V. cholerae C6706str2 ΔVC1188 pZND95 This study ZDC587 V. cholerae C6706str2 ΔVC1280 pZND95 This study ZDC588 V. cholerae C6706str2 ΔVC1323 pZND95 This study ZDC589 V. cholerae C6706str2 ΔVC1325 pZND95 This study ZDC590 V. cholerae C6706str2 ΔVC1327 pZND95 This study ZDC591 V. cholerae C6706str2 ΔVC1328 pZND95 This study ZDC592 V. cholerae C6706str2 ΔVC1361 pZND95 This study ZDC593 V. cholerae C6706str2 ΔVC1590 pZND95 This study ZDC594 V. cholerae C6706str2 ΔVC1594 pZND95 This study ZDC595 V. cholerae C6706str2 ΔVC1595 pZND95 This study ZDC596 V. cholerae C6706str2 ΔVC1596 pZND95 This study ZDC597 V. cholerae C6706str2 ΔVC2647 pZND95 This study ZDC598 V. cholerae C6706str2 ΔVCA0935 pZND95 This study ZDC651 V. cholerae C6706str2 ΔVC0704 pZND122, pZND95 This study ZDC653 V. cholerae C6706str2 ΔVC1280 pZND122, pZND95 This study ZDC655 V. cholerae C6706str2 ΔVC1323 pZND122, pZND95 This study ZDC657 V. cholerae C6706str2 ΔVC1595 pZND122, pZND95 This study ZDC659 V. cholerae C6706str2 ΔVCA0935 pZND122, pZND95 This study ZDC661 V. cholerae C6706str2 ΔVC1327 pZND122, pZND95 This study ZDC570 V. cholerae C6706str2 ΔvpsL ΔluxO pZND122 This study
CW2034 V. cholerae C6706str2 ΔvpsL None (Waters et al., 2008)
CW2035 V. cholerae C6706str2 ΔvpsL luxO-D47E None (Waters et al., 2008)

55
CW2036 V. cholerae C6706str2 ΔvpsL ΔhapR None (Waters et al., 2008)
CW2037 V. cholerae C6706str2 ΔvpsL ΔluxO None (Waters et al., 2008)
MM1162 E. coli S17-1λpir Wild type pKAS32-ΔhapR (Miller et al. 2002)
ZDE92 E. coli S17-1λpir Wild type pZND46 This study
WN778 V. cholerae C6706str2 luxO-D47E None WL Ng, unpublished
BH2126 V. cholerae C6706str2 luxO-D47E Δqrr1-4 None (Hammer and Bassler, 2007)
WN780 V. cholerae C6706str2 Δqrr1-4 None WL Ng, unpublished
SLS501 V. cholerae C6706str2 Δqrr1-4 ΔhapR None (Svenningsen et al., 2008)
Table 3.3 Bacterial strains used in this study

56
VC0583 hemagglutinin/protease regulatory protein, authentic frameshift Regulatory functions
VC0704
spermidine/putrescine ABC transporter, periplasmic spermidine/putrescine-binding protein, putative
Transport and binding proteins
VC0811 hypothetical protein Unknown VC0975 conserved hypothetical protein Hypothetical proteins VC0976 conserved hypothetical protein Hypothetical proteins VC1188 malate oxidoreductase Energy metabolism VC1280 hypothetical protein Unknown VC1323 hypothetical protein Unknown
VC1325 galactoside ABC transporter, periplasmic D-galactose/D-glucose-binding protein
Transport and binding proteins
VC1327 galactoside ABC transporter, ATP-binding protein Transport and binding proteins
VC1328 galactoside ABC transporter, permease protein Transport and binding proteins
VC1361 amino acid ABC transporter, permease protein Transport and binding proteins
VC1590 acetolactate synthase, catabolic Amino acid biosynthesis VC1594 aldose 1-epimerase Energy metabolism VC1595 galactokinase Energy metabolism VC1596 galactose-1-phosphate uridylyltransferase Energy metabolism VC2647 conserved hypothetical protein Hypothetical proteins VCA0935 hypothetical protein Unknown
Table 3.3 Genes studied in the microarray screen

57
CHAPTER 4:
CQSA AND CQSS REGULATION IS POST-TRANSCRIPTIONAL

58
Introduction
Quorum sensing is a cell-to-cell communication mechanism used by bacteria to
determine their population density and regulate gene expression accordingly. Vibrio
cholerae is a marine bacterium and human pathogen that contains two QS pathways
which converge into a shared phosphorelay to control the QS response. One of these
QS systems, the CqsA/S system, contains a positive feedback loop controlled by indirect
repression from the Qrr sRNAs onto CqsA/S. Here, we investigate CqsA/S repression
and show that the mechanism responsible for CqsA/S repression is post-transcriptional.
Additionally, we identify regions within the cqsA gene that could serve as potential sites
for post-transcriptional regulation.
Results
cqsA and cqsS are post-transcriptionally regulated
The Qrr sRNAs indirectly regulate the level of cqsA and cqsS RNAs in V.
cholerae. Here, we further investigate cqsA and cqsS gene expression by constructing
gene fusions to dissect the specific mechanism of regulation. We first aimed to
determine if the cqsA and cqsS mRNAs are regulated at the transcriptional or post-
transcriptional level. We constructed plasmids containing IPTG-inducible cqsA-FLAG
and cqsS-FLAG by replacing their endogenous promoters with the E. coli lacZYA
promoter (Figure 5.1). We transformed these plasmids into three V. cholerae strains that
differed in Qrr sRNA expression: a ΔvpsL strain, a ΔvpsL luxO-D47E strain, and a ΔvpsL
ΔluxO strain harboring a plasmid carrying Ptet-qrr4. Addition of aTc to the ΔvpsL ΔluxO
strain harboring Ptet-qrr4 causes qrr4 induction, allowing this strain to exhibit either a
locked LCD (induced) or locked HCD (uninduced) phenotype. cqsA-FLAG or cqsS-FLAG

59
expression was induced with IPTG in these strains, and CqsA-FLAG or CqsS-FLAG was
measured at three time points during growth from exponential to stationary phase. The
results show that both cqsA and cqsS expression are Qrr regulated even when driven by
the lac promoter, suggesting that regulation is post-transcriptional (Figures 4.2 and 4.3).
CqsA-FLAG and CqsS-FLAG levels are similar between ΔvpsL and ΔvpsL ΔluxO strains
when qrr4 is not induced. Conversely, CqsA-FLAG and CqsS-FLAG levels exhibit low
expression in the ΔvpsL luxO-D47E and ΔvpsL ΔluxO strains when qrr4 is induced.
To further investigate if cqsA regulation is post-transcriptional, we constructed
plasmids containing promoter fusions and transcriptional fusions linking the cqsA
promoter and 5’UTR with the gfp open reading frame (Figure 4.4). These plasmids were
introduced into a V. cholerae ΔvpsL ΔluxO strain harboring a plasmid with Ptet-qrr4. The
results show that gfp expression does not change when qrr4 is induced, suggesting that
Qrr4 does not regulate the cqsA promoter or 5’UTR. Again, this is consistent with post-
transcriptional regulation.
Screens for a cqsA regulator
We attempted to screen for regulators of cqsA and cqsS mRNA, however,
numerous problems arose. Fluorescent protein signals from V. cholerae are significantly
less bright compared to species such as V. harveyi and E. coli (Knut Drescher,
unpublished data). Although fluorescent constructs have been used successfully for
screening for V. cholerae mutants, such as Pqrr4-gfp, these constructs are generally
transcriptional fusions that are driven by strong promoters. Thus, all attempts to
construct cqsA and cqsS promoter, transcriptional, and translational fusions to
fluorescent proteins in V. cholerae failed to yield signal on FACS and in 96-well plate

60
readers, despite using the brightest available fluorescent proteins. Therefore, screens for
cqsA or cqsS expression using fluorescent proteins is not possible in V. cholerae.
Screening in E. coli relies on induced cqsA expression because cqsA is not expressed
from its endogenous promoter in E. coli. Additionally, the dynamic range for cqsA
expression in V. cholerae is dramatically lower when cqsA is not driven by its
endogenous promoter, which limits the usefulness of a Plac-cqsA-gfp construct in both E.
coli and V. cholerae when a large range of expression levels is needed to identify
mutants exhibiting altered expression.
For these reasons, we devised a screen that takes into account the above
experimental constraints. We constructed a plasmid containing an IPTG-inducible Plac-
cqsA-gfp fusion, and transformed it into E. coli harboring aTc-inducible V. cholerae
genomic fragments. Our rationale was that if a V. cholerae genomic fragment contained
a gene that regulates cqsA post-transcriptionally, it would either increase or decrease
GFP expression, depending on whether it activated or repressed cqsA. Despite repeated
attempts, we consistently found that the GFP expression among preliminary hits was
heterogeneous and not repeatable.
Because direct screening for cqsA regulation proved difficult, we additionally
conducted a candidate screen to attempt to identify a cqsA regulator based on the
following criteria. First, we reasoned that cqsA is likely regulated post-transcriptionally,
and second, we demonstrated that cqsA expression is decreased in an Δhfq mutant
(data not shown), suggesting a sRNA as a candidate regulator. Third, if only one
intermediate exists between the Qrr sRNAs and cqsA, the sRNA intermediate would
need to exhibit binding specificity to both qrr4 and cqsA. Fourth, if the QS control of cqsA
levels is linked to the growth phase regulation of cqsA (see chapter 2), we would expect
to identify an intermediate whose own expression changes during the transition from

61
exponential to stationary phase growth. Fifth, research from other groups has suggested
that CRP may indirectly regulate cqsA expression post-transcriptionally (Liang et al.,
2008), and we therefore narrowed our search to sRNAs that are regulated by glucose.
We identified two sRNA regulators that fit all of these criteria. Spot42 and VSsrna24 are
both sRNAs that regulate mRNAs post-transcriptionally, appear to be QS- and CRP-
controlled, and differ in concentration during the transition from exponential to stationary
phase growth. We investigated if these RNAs could control cqsA by expressing aTc-
inducible promoter fusions to Spot42 and VSsRNA24 and measuring cqsA mRNA levels
in V. cholerae QS mutants (data not shown). Our results show that neither Spot42 nor
VSsRNA24 affect cqsA RNA expression levels.
Determining the site of cqsA regulation.
Because we were not able to identify a gene responsible for regulating cqsA, we
attempted to identify a location on the cqsA RNA that was necessary and sufficient for
Qrr-based regulation. Our rationale was that if we could identify a sequence in cqsA
necessary and sufficient for regulation, would could use BLAST analysis of the V.
cholerae genome to identify complementary putative sRNA regulator sequences. We
first constructed IPTG-inducible gene fusions that replaced the cqsA and cqsS 5’UTRs
with the E. coli cmR (chloramphenicol resistance gene) 5’UTR to determine if the cqsA/S
5’UTR, a common target for sRNA regulation, is recognized for regulation. These
constructs failed to yield any signal when examined by Western blotting and Northern
blotting (data not shown). We also constructed IPTG-inducible gene fusions that
replaced the cqsA and cqsS ORF with the gfp ORF. Again, these fusions did not yield
consistent gfp expression by Northern and Western blotting (data not shown). We next
took a more nuanced approach to determine the site of Qrr-based regulation. We

62
constructed plasmids containing sequential truncations of the N- and C-termini of cqsA in
increments of 150bp to determine which region is necessary for Qrr regulation of cqsA.
These plasmids were introduced into a V. cholerae ΔvpsL ΔluxO ΔcqsA strain harboring
a plasmid with Ptet-qrr4 (Figures 4.5 and 4.6). Our rationale was that if a mutant failed to
show cqsA repression by Qrr4, then the deleted region is necessary for cqsA regulation.
All C-terminal truncation mutants showed substantially decreased basal expression
when compared to wild-type expression, but none showed substantial differences in
cqsA levels when qrr4 was induced. The N-terminal truncation mutant series showed a
different pattern of expression than for the C-terminal truncation mutants. The 150bp N-
terminal truncation showed no cqsA expression, even when Qrr4 expression was not
induced. The only mutant that was not regulated by Qrr4 was the 900bp N-terminal
truncation (Figure 4.6).
Discussion
The autoinducer-receptor pair cqsA and cqsS are regulated at the mRNA level by
QS through an indirect mechanism. We proposed that this regulation could occur
through a transcriptional method via regulation of the production of the mRNAs, or could
occur post-transcriptionally via the process of degrading existing mRNAs. We examined
this question by constructing gene fusions that separated the promoters and the open
reading frames of these genes and studying them independently. We found that both
cqsA and cqsS are regulated post-transcriptionally. This mode of feedback is contrary to
most known QS systems, which use transcriptional regulation to impose positive
feedback in the QS system. However, this mode of feedback is similar in architecture to
the luxMN system in V. harveyi that uses direct Qrr regulation of the luxMN transcript to
generate positive feedback. The cqsA/S post-transcriptional regulatory mechanism could

63
be either an RNA binding protein or an RNA regulator. sRNA-sRNA interactions are not
widely reported in bacteria, but have been observed often in eukaryotes in the form of
miRNA sponges (Ebert et al., 2007).
There are many possible reasons why V. cholerae uses post-transcriptional
regulation over transcriptional for cqsA/S. First, post-transcriptional regulation may show
faster dynamics than transcriptional regulation. Although the timing for Qrr feedback onto
cqsA is similar to what is expected for transcriptional regulation, the intermediate
between the Qrr sRNAs and cqsA/S may receive additional inputs that can confer a
rapid response to changing environmental conditions. Second, the V. cholerae QS
response is opposite from most pathogens in that virulence factor and biofilm production
are activated at LCD. By activating cqsA/S through a post-transcriptional mechanism,
these genes are constantly being transcribed and therefore their products are primed to
rapidly accumulate in concentration in the event that the Qrr sRNAs are depleted. Third,
since sRNAs play a large role in the QS responses of V. cholerae and are responsible
for regulating multiple QS genes, V. cholerae may have evolved use of sRNAs as the
preferred post-transcriptional regulation mechanism as a residual feature of V.
cholerae’s evolutionary history.
We also examined the cqsA transcript to determine which regions of the
transcript are necessary for the post-transcriptional repression. We discovered though
gene fusions and truncations of cqsA that the transcript is sensitive to modification.
Deletions of, or fusions to, the 3’ end of the transcript dramatically lower the basal level
of cqsA expression. Because of these observations, we hypothesize that that the 3’ end
of the cqsA transcript may be necessary for stability of the transcript. One possibility is
that this region of the transcript is somehow tied to regulation, for instance, this region

64
may be necessary for stabilizing the transcript, and modifications to the secondary
structure from a regulator serve to prime the transcript for degradation by exonucleases.

65
A.
B.
Figure 4.1 cqsA and cqsS fusion constructs
A. cqsA and B. cqsS fusions were constructed on the pEVS143 plasmid backbone.
Endogenous promoters contained ~500bp of sequence upstream of the ORF. The lac
promoters consisted of tac promoters from pEVS143 modified to the lac sequence. All
constructs contained the first rho-independent terminator from the rrnB gene of E. coli to
stop transcription.

66
Figure 4.2 Post-transcriptional regulation of cqsA
CqsA-FLAG expression was measured by Western blotting. V. cholerae mutants
harboring IPTG-inducible cqsA-FLAG fusions were induced with 100uM IPTG and grown
in shaking flasks. 50ng/mL aTc was added to one flask to induce Qrr4 production.
Aliquots were taken at OD600 = 0.2, 0.8, and 1.4 and processed for Western blotting.

67
Figure 4.3 Post-transcriptional regulation of cqsS
CqsS-FLAG expression was measured by Western blotting. V. cholerae mutants
harboring IPTG-inducible cqsS-FLAG fusions were induced with 100uM IPTG and grown
in shaking flasks. 50ng/mL aTc was added to one flask to induce Qrr4 production.
Aliquots were taken at OD600 = 0.2, 0.8, and 1.4 and processed for Western blotting.

68
Figure 4.4 cqsA promoter is not regulated by Qrr4
cqsA expression was measured by Northern blotting. A ΔvpsL ΔluxO ΔcqsA V. cholerae
strain harboring a plasmid with Ptet-qrr4 and a gfp-FLAG gene controlled by the cqsA
promoter was induced with 100uM IPTG and grown in shaking flasks. 50ng/mL aTc was
added to flasks to induce Qrr4 production. Aliquots were taken at OD600 = 1.0 and
processed for Northern blotting.

69
Figure 4.5 C-terminal cqsA truncations
cqsA expression was measured by Northern blotting. Full-length cqsA and cqsA C-
terminal truncations were expressed from the endogenous promoter on plasmids in a V.
cholerae ΔvpsL ΔluxO ΔcqsA strain harboring a plasmid with Ptet-qrr4. All samples were
measured at OD=1.0. “+” indicates Qrr4 induction with with 50ng/mL aTc, “-“ indicates no
Qrr4 induction. “Δ” indicates the bases of the cqsA ORF that are deleted.

70
Figure 4.6 N-terminal cqsA truncations
cqsA expression was measured by Northern blotting. Full-length cqsA and cqsA N-
terminal truncations were expressed from the endogenous promoter on plasmids in a V.
cholerae ΔvpsL ΔluxO ΔcqsA strain harboring a plasmid with Ptet-qrr4. All samples were
measured at OD=1.0. “+” indicates Qrr4 induction with with 50ng/mL aTc, “-“ indicates no
Qrr4 induction. “Δ” indicates the bases of the cqsA ORF that are deleted.

71
Materials and Methods
Bacterial strains and media
V. cholerae El Tor C6706str2 and isogenic mutant strains were grown at 30°C
and E. coli S17-1λpir (de Lorenzo and Timmis, 1994) strains were grown at 37°C in
Luria-Bertani (LB) medium. Liquid cultures were grown in flasks or test tubes with
shaking for aeration. Strains used in this study are noted in Table 4.3. Antibiotics were
used at the following concentrations: chloramphenicol 10 µg/mL, ampicillin 100ug/mL,
kanamycin 100ug/mL. Plasmids were electroporated into electrocompetent E. coli S17-
1λpir by the MicroPulser (Bio-Rad).
DNA manipulations and plasmids
Plasmid construction was performed by standard methods. Polymerase chain
reactions (PCR) were performed using the iProof DNA polymerase (Bio-Rad), and
restriction digestions and ligations were performed using restriction endonucleases (New
England Biolabs) and T4 DNA ligase (NEB). Phosphorylation of unphosphorylated DNA
ends was accomplished by T4 polynucleotide kinase (NEB) according to the
manufacturer’s guidelines.
Plasmid pZND109 was constructed via semi-blunt cloning by PCR-amplifying
cqsA from V. cholerae gDNA and PCR-amplifying the pEVS143 backbone vector,
digestion of amplicons with KpnI-HF, phosphorylation of DNA ends, and ligation.
pZND116 was created by PCR-amplifying the cmR resistance gene from pJS1194 and
subcloning and replacing the kanR gene this gene on pZND109. PCR amplicons were
digested with SalI-HF and AvrII and ligated together. Plasmid pZND111 was generated
by semi-blunt cloning via PCR-amplifying gfp from pEVS143 as the insert, and PCR-
amplifying a different region of pEVS143 as the vector backbone. The relevant cqsA

72
fragments were included in the PCR primers. Amplicons were digested with KpnI-HF,
phosphorylated, and ligated. pZND117 was constructed identically as pZND116 except
pZND111 was used in place of pZND109. Plasmid pZND120 was constructed via semi-
blunt cloning by PCR-amplifying cqsS from V. cholerae gDNA and amplifying the
pZND116 backbone as the cloning vector. Amplicons were digested with KpnI-HF,
phosphorylated, and ligated together. Plasmid pZND121 was generated via semi-blunt
cloning by PCR amplifying the gfp ORF from pEVS143 and the cqsS 5’UTR from V.
cholerae gDNA. These amplicons were combined using SOEing PCR to create the
cloning insert, and the cloning vector was generated by PCR amplifying the pZND111
backbone. The amplicons were restriction digested with KpnI-HF, phosphorylated, and
ligated together. Plasmid pZND129 was generated by PCR-amplifying 522bp of the cqsA
promoter from V. cholerae gDNA and superfolderGFP from pNUT173. These amplicons
were combined using SOEing PCR. The vector was PCR-amplified from the pZND116
plasmid backbone. The PCR fragments were restriction digested with EcoRI-HF and
KpnI-HF and subsequently ligated. Plasmid pZND130 was constructed in an identical
manner as pZND129, except the pZND130 insert primers contained sequences for a
superfolderGFP 5’UTR rather than a cqsA 5’UTR.
Plasmid pZND45 was constructed to delete a 75bp segment of the V. cholerae
genome comprising the RNA Polymerase binding region for cqsA. The 800bp regions
upstream and downstream of “TAA AAA TAA TTT TTC CCA GAT GAC AGA CGT TTT
TAA TCA CGC AAT ATA TCA CTC GTC AGC TGG CGT TAA ATT TTT” were PCR-
amplified and combined using SOEing PCR. The PCR amplicon and pKAS32 were
restriction digested with NotI-HF and EcoRI-HF and ligated together.
Plasmids pZND145--pZND154 and pZND157 were all constructed using a similar
methodology. Primers were used to PCR-amplify the backbone of pZND95, but were

73
placed to allow for truncations of the cqsA ORF in multiples of 150bp. Either the forward
or reverse primer was engineered to contain a 5’ phosphate to allow for blunt-end
ligation immediately after PCR amplification. All plasmids were transformed into E. coli
S17-1λpir, and V. cholerae-harbored plasmids were mated to V. cholerae strains as
described (Skorupski and Taylor, 1996).
Northern Blots
cqsA riboprobes were synthesized by first PCR-amplifying V. cholerae El Tor
C6706str2 gDNA template with primers in Table 4.2, and then performing T7 in vitro
transcription on the PCR amplicon (Ambion) with 32P-α-UTP. qrr4, spot42, and VSsrna24
oligoprobes were synthesized by end-labeling primers in Table 4.2 with 32P-γ-ATP using
T4 PNK (New England Biolabs). Riboprobes and oligoprobes were purified by Illustra
Microspin columns according to the manufacturer’s guidelines (GE Healthcare). For
Northern blot experiments, overnight cultures were back-diluted 1:1000 into fresh LB and
shaken in test tubes at 30°C, grown to OD=1.0. 5 OD units of culture were added to 20%
RNA stop solution (95% ethanol, 5% phenol), mixed by inversion, and frozen in liquid
nitrogen. Samples were stored at -80°C until processing the following day. To isolate
RNA from the samples, samples were thawed at room temperature and centrifuged at
5000x g for 10 minutes at 4°C. Supernatants were discarded and pellets were processed
to extract RNA following the Trizol method (Tu and Bassler, 2007). 10ug of total RNA
was resolved on 6% polyacrylamide (PAA, 7M urea) at 300V for 2 hours. RNA was
transferred to Amersham Hybond-XL nylon membranes (Amersham Biosciences,
Piscataway, NJ) for 1 hour at 50V and at 4°C. For hybridization, membranes were
incubated overnight at 70°C for cqsA riboprobes and at 42°C for qrr4 oligoprobes in

74
15mL Rapid-hyb buffer (GE Healthcare, Piscataway, NJ). Following overnight incubation,
membranes were washed three times in SSC buffer + 0.1%SDS (5X, 1X, and 0.5X at
42°C for oligoprobes; 2X, 1X, and 0.5X at 70°C for riboprobes). Blots were imaged for 20
hours unless otherwise noted on phosphorimager screens and scanned on a Typhoon
9410 (GE Healthcare).
Western blots
Overnight cultures were back-diluted 1:1000 into fresh LB and shaken in flasks at
30°C, and OD measurements were conducted periodically. Cell cultures were isolated at
OD = 0.2, 0.8, and 1.4, and 1 OD unit of culture was immediately centrifuged at
16,000xg for 1 minute, the supernatant was removed, and pellets were stored at -80°C
until further processing. To prepare protein from pellets, pellets were dissolved in Bug
Buster (Millipore) and incubated at room temperature for 20 minutes. Samples prepared
in Bug Buster solution were mixed with 0.5% SDS. The Bug Buster+SDS mix was boiled
for 3 minutes before being run on an SDS-PAGE gel. 10µl of each sample was
electrophoresed at 150V for 2 hours on 15-well 4-15% gradient SDS-PAGE gels (Bio-
Rad). Experiments testing for CqsA-FLAG were wet blotted onto nitrocellulose
membranes, and experiments testing for CqsS-FLAG were wet blotted onto PVDF
membranes pre-soaked in methanol. After blotting, membranes were cut in half to allow
the RNA polymerase β’ subunit control and FLAG epitopes to be probed independently.
After blotting, membranes were blocked in TBST+5% dry milk for 1 hour with shaking at
room temperature. Subsequently, α-FLAG-HRP antibody (Sigma-Aldrich) was diluted
1:5000 in TBST and incubated against the FLAG membrane for 2 hours. Simultaneously,
α-RNA polymerase β’ subunit antibody (Abcam) was diluted 1:100,000 in TBST and
incubated against the RNA polymerase β’ subunit membrane for 1 hour. The

75
membranes testing for α-RNA polymerase β’ subunit were rinsed with TBST and
incubated with α-mouse-HRP for 1 hour. After this incubation, both membranes
containing the FLAG and α-RNA polymerase β’ subunit were rinsed with TBST and
incubated with Amersham EL Prime Western Blotting Detection Reagent according to
the manufacturer’s guidelines. Membranes treated with the chemiluminescent reagents
were visualized via film exposure for 5 minutes.

76
Plasmid name Description Source pEVS143 Empty vector (Dunn et al., 2006) pJS1194 Empty vector J. Schaffer, unpublished pKAS32 Empty vector (Skorupski and Taylor, 1996) pZND109 pEVS143 with Plac-cqsA-FLAG This study pZND111 pEVS143 with Plac-cqsA5'UTR-gfp-FLAG This study pZND116 pEVS143 with Plac-cqsA-FLAG This study pZND117 pEVS143 with Plac-cqsA5'UTR-gfp-FLAG This study pZND120 pEVS143 with Plac-cqsS-FLAG This study pZND121 pEVS143 with Plac-cqsS5'UTR-gfp-FLAG This study pZND122 pASK75 with Ptet-qrr4 and oriT This study pZND123 pEVS143 with Plac-CmR5'UTR-cqsA-FLAG This study pZND124 pEVS143 with Plac-CmR5'UTR-cqsS-FLAG This study pZND129 pEVS143 with PcqsA-cqsA5'UTR-gfp-3xFLAG This study pZND130 pEVS143 with PcqsA-gfp5'UTR-gfp-3xFLAG This study pZND143 pEVS143 with Ptac-spot42 K. Papenfort, unpublished pZND144 pEVS143 with Ptac-VSsrna24 K. Papenfort, unpublished pZND145 pZND95 cqsA C-terminal truncation (150bp) This study pZND146 pZND95 cqsA C-terminal truncation (300bp) This study pZND147 pZND95 cqsA C-terminal truncation (450bp) This study pZND148 pZND95 cqsA C-terminal truncation (600bp) This study pZND149 pZND95 cqsA C-terminal truncation (750bp) This study pZND150 pZND95 cqsA C-terminal truncation (1050bp) This study pZND151 pZND95 cqsA N-terminal truncation (150bp) This study pZND152 pZND95 cqsA N-terminal truncation (300bp) This study pZND153 pZND95 cqsA N-terminal truncation (450bp) This study pZND154 pZND95 cqsA N-terminal truncation (600bp) This study pZND157 pZND95 cqsA N-terminal truncation (900bp) This study pZND45 pKAS32-ΔPcqsA This study pZND95 pJS1194 with cqsA-FLAG This study
Table 4.1 Plasmids used in this study

77
Primer name Sequence Use 468 GTC AGC TGG CGT TAA ATT TTT TAT AAC TAG G pZND109 (insert)
463
AAA AAG GTA CCA AAA CGA AAG GCC CAG TCT TTC GAC TGA GCC TTT CGT TTT ACT TGT CGT CAT CGT CTT TGT AGT CAC GAA AAT AAA AAT CAC CGT AGT TGA CCG pZND109 (insert)
469 AAA AAG GTA CCG ATC CGG TGA TTG ATT GAG CAA GC
pZND109, pZND111, pZND121 (vector)
464
GTG CTC AAC ATA TTG TTA TCC GCT CAC AAT GTA AAT TGT TAT CCG CTC ACA ACA GCT CAT TTC AGA ATA TTT GCC AGA ACC
pZND109, pZND111, pZND120, pZND121 (vector)
523 AAA AAA GTC GAC GAA GAT GCG TGA TCT GAT CCT TCA ACT C pZND116 (insert)
524 AAA AAA CCT AGG CGT TGT GTC TCA AAA TCT CTG ATG TTA CAT TGC pZND116 (insert)
525 AAA AAA CCT AGG ACT CGC TAC GCT CGG TCG TTC GAC T pZND116 (vector) 526 AAA AAA GTC GAC ATC ACG CAT CTT CCC GAC AAC GCA GA pZND116 (vector)
466 GTC AGC TGG CGT TAA ATT TTT TAT AAC TAG GAT ATA TTG CGA TGG CTA GCA AAG GAG AAG AAC TCT TC pZND111 (insert)
467
AAA AAG GTA CCA AAA CGA AAG GCC CAG TCT TTC GAC TGA GCC TTT CGT TTT ACT TGT CGT CAT CGT CTT TGT AGT CGT TGT ACA GTT CAT CCA TGC CAT GTG TAA TC
pZND111, pZND121 (insert)
501 GAT GAA GGT TTT GGC AGT TTG GAT CCG pZND120 (insert) 519 GTG CTC AAC ATA TTG TTA TCC GCT CAC A pZND120 (vector)
474
AAA AAG GTA CCA AAA CGA AAG GCC CAG TCT TTC GAC TGA GCC TTT CGT TTT ACT TGT CGT CAT CGT CTT TGT AGT CCA CCC AAG CTG CCA CTT TAT TTA GC pZND120 (insert)
522 GAT GAA GGT TTT GGC AGT TTG GAT CCG G pZND121 (insert)
502 TCC TTT GCT AGC CAT GCT CAC TAT CAC TAC CGT TGC ATT CTC pZND121 (insert)
503 GTA GTG ATA GTG AGC ATG GCT AGC AAA GGA GAA GAA CTC TTC pZND121 (insert)
527 TTT TGA ATT CTT GCG CAG CCC GAC CCG ATT CT pZND129, pZND130 (insert)
531 TTC TCC TTT GCT CAT CGC AAT ATA TCC TAG TTA TAA AAA ATT TAA CGC CA pZND129 (insert)
532 CTA GGA TAT ATT GCG ATG AGC AAA GGA GAA GAA CTT TTC ACT GG pZND129 (insert)
533
AAA AGG TAC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTA TTT ATC GTC ATC TTT GTA GTC GAT ATC ATG ATC TTT ATA ATC ACC GTC ATG GTC TTT GTA GTC TTT GTA GAG CTC ATC CAT GCC ATG TGT AAT C
pZND129, pZND130 (insert)
529 TTT TGG TAC CGA TCC GGT GAT TGA TTG AGC pZND129, pZND130 (vector)
530 TTT TGA ATT CCA GAA CCG TTA TGA TGT CGG CGC pZND129, pZND130 (vector)
534 ATC TCC TTA ACT AGG GAG TGA TAT ATT GCG TGA TTA AAA ACG TCT GTC pZND130 (insert)
535 CGC AAT ATA TCA CTC CCT AGT TAA GGA GAT ATA CAT ATG AGC AAA GGA pZND130 (insert)
271 AAA AGC GGC CGC CTA TGC GAC TAT CTC GCG ATT C pZND45 (insert)

78
272 CAA TAT ATC CTA GTT ATA TTA GCA ACT TAG AAT AAT TAA TAA TTC TCG ATG pZND45 (insert)
273 TT ATT CTA AGT TGC TAA TAT AAC TAG GAT ATA TTG CGA TGA pZND45 (insert)
274 AAA AGA ATT CGG AAC GCA GCG ATT CAC TTC AT pZND45 (insert)
576 GTG TTT GGC TCA GTA TTC TGC CGC cqsA 3' region riboprobe
577 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG ACG AAA ATA AAA ATC ACC GTA GTT GAC CGC
cqsA 3' region riboprobe
567 GTC AGC TGG CGT TAA ATT TTT TAT AAC TAG G cqsA 5' region riboprobe
568 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG CTT GTT TAC CCA ATA CAA GGT GTT TAC CG
cqsA 5' region riboprobe
541 CTG TCC GTG GAG AGG GTG AA superfolderGFP riboprobe
542 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG ATC CGG ATA ACG GGA AAA GC
superfolderGFP riboprobe
539 CCA GCC CAA TAC GAA TGT TT cqsA riboprobe
540 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG CAA TGA TCC CAG GAC CAT GAC G cqsA riboprobe
Table 4.2 Primers used in this study

79
Strain name Organism Genotype Plasmid Source
CW2034 V. cholerae C6706str2 ΔvpsL None Waters et. al, 2008
CW2035 V. cholerae C6706str2 ΔvpsL luxO-D47E None Waters et. al, 2008
CW2037 V. cholerae C6706str2 ΔvpsL ΔluxO None Waters et. al, 2008
ZDC621 V. cholerae C6706str2 ΔvpsL pZND116 This study ZDC623 V. cholerae C6706str2 ΔvpsL pZND120 This study ZDC631 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND116 This study ZDC633 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND120 This study ZDC641 V. cholerae C6706str2 ΔvpsL ΔluxO pZND122, pZND116 This study ZDC643 V. cholerae C6706str2 ΔvpsL ΔluxO pZND122, pZND120 This study ZDC702 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA None This study ZDC757 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND95, pZND122 This study ZDC775 V. cholerae C6706str2 ΔvpsL pZND143 This study ZDC777 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND143 This study ZDC779 V. cholerae C6706str2 ΔvpsL ΔluxO pZND143, pZND122 This study ZDC811 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND122, pZND129 This study ZDC823 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND122, pZND130 This study ZDC839 V. cholerae C6706str2 ΔvpsL pZND144 This study ZDC841 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND144 This study ZDC845 V. cholerae C6706str2 ΔvpsL ΔluxO pZND144, pZND122 This study ZDC853 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND145 This study ZDC855 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND146 This study ZDC857 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND147 This study ZDC859 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND148 This study ZDC861 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND149 This study ZDC863 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND150 This study ZDC865 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND151 This study ZDC869 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND152 This study ZDC871 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND153 This study ZDC873 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND154 This study ZDC875 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND157 This study ZDE244 E. coli S17-1λpir Wild type pZND95 This study ZDE362 E. coli S17-1λpir Wild type pZND122 This study ZDE429 E. coli S17-1λpir Wild type pZND116 This study ZDE433 E. coli S17-1λpir Wild type pZND120 This study ZDE497 E. coli S17-1λpir Wild type pZND129 This study ZDE499 E. coli S17-1λpir Wild type pZND130 This study

80
ZDE540 E. coli S17-1λpir Wild type pZND145 This study ZDE542 E. coli S17-1λpir Wild type pZND146 This study ZDE544 E. coli S17-1λpir Wild type pZND147 This study ZDE546 E. coli S17-1λpir Wild type pZND148 This study ZDE548 E. coli S17-1λpir Wild type pZND149 This study ZDE550 E. coli S17-1λpir Wild type pZND150 This study ZDE552 E. coli S17-1λpir Wild type pZND151 This study ZDE589 E. coli S17-1λpir Wild type pZND152 This study ZDE590 E. coli S17-1λpir Wild type pZND153 This study ZDE592 E. coli S17-1λpir Wild type pZND154 This study ZDE90 E. coli S17-1λpir Wild type pZND45 This study
Table 4.3 Bacterial strains used in this study

81
CHAPTER 5:
EXAMINATION OF CQSA GROWTH PHASE REGULATION

82
Introduction
Quorum sensing is a communication process used by bacteria to assess their
population size and regulate the expression of genes involved in group behaviors. The
QS system of the human pathogen V. cholerae operates through the production of
autoinducers by the synthases CqsA and LuxS, and detection of autoinducers by the
receptors CqsS and LuxPQ, respectively. Previous chapters have shown that the
autoinducer synthase gene cqsA is not transcribed in E. coli under the native cqsA
promoter, suggesting that an activator of cqsA may exist in V. cholerae, but not in E. coli.
Here, we screen for activators of cqsA and examine candidates for their roles in V.
cholerae QS. This screen identifies multiple genes with potential roles in regulation of the
Qrr sRNAs.
Results Screens for a cqsA transcriptional activator
We have previously shown that cqsA is not expressed from its endogenous
promoter in E. coli. However, cqsA expression can be detected in E. coli when cqsA is
expressed from an IPTG-inducible promoter. We hypothesized that this discrepancy
could be explained if a transcriptional activator of cqsA exists in V. cholerae, but not in E.
coli. In order to test this hypothesis, we devised two screens to isolate a cqsA
transcriptional activator.
In the first screen, we constructed an E. coli strain harboring a PcqsA-gfp
transcriptional fusion. The cells carrying this fusion were dark since this promoter is not
expressed in E. coli. We next introduced a V. cholerae genomic library under an aTc-
inducible Ptet promoter into this E. coli strain. Our rationale was that if a V. cholerae
genomic fragment contained an activator of the cqsA promoter, GFP expression would

83
be induced and the cell would be bright. After multiple rounds of FACS sorting and
measurements on a 96-well plate reader, eleven clones were isolated and retested, but
ultimately none consistently exhibited GFP expression above background levels.
In the second screen, we constructed a V. cholerae Tn5 transposon library
carrying a Pqrr4-gfp promoter fusion. We conducted the screen in a V. cholerae ΔluxQ
background to eliminate qrr4 activation that would stem from the AI-2 signaling pathway.
We used a Pqrr4-gfp fusion rather than the PcqsA-gfp fusion because the PcqsA-gfp
fusion does not fluoresce in V. cholerae. Our rationale was that clones expressing cqsA
would be dark because these clones would be in HCD mode and thus, repress the qrr4
promoter. Transposon mutants deficient in cqsA activation caused by transposon
insertion would, by contrast, be in LCD mode, and the qrr4 promoter would be activated,
causing GFP to be expressed. We first confirmed that the pattern of GFP expression
from Pqrr4-gfp matched our expectations of high GFP using a cqsA null allele (Figure
5.1) Subsequently, we screened the V. cholerae Tn5 library and isolated seven targets
from the initial screen, and reconfirmed these clones using a 96-well plate reader assay
and FACS. We found that four of the seven transposon mutants showed a fluorescence
signal similar to that of a ΔluxQ ΔcqsA control strain (Figure 5.2), and we sequenced the
insertion sites in these mutants.
The transposon insertions were located inside three ORFs: VCA0033, VC0122,
and VC1926 (two separate insertions). VCA0033 is a hypothetical gene that has been
suggested to show homology to phoA (Majumdar et al., 2005). VC0122 encodes one of
the two V. cholerae adenylate cyclase genes, cya. VC1926 encodes the gene dctD, a
NtrC-class activator and two-component system response regulator. NtrC-class

84
activators bind far upstream of their target promoters and activate transcription through
DNA looping and ATPase activity, and they interact with the alternative sigma factor σ54.
In previous chapters, we have demonstrated that the Qrr sRNAs repress cqsA
through a feedback loop, and therefore mutations that alter Qrr levels can affect cqsA
mRNA levels. Because our experimental system relied on measuring signal from the
qrr4 promoter and not from cqsA itself, we further investigated the mutants from the
present screen by directly measuring RNA levels of both cqsA and qrr4 at exponential
and stationary phases (Figure 5.3). We found that at exponential phase (OD600 = 0.1),
the VCA0033 and VC1926 mutants contained higher cqsA mRNA levels than wild-type,
whereas the VC0122 mutant showed cqsA mRNA levels identical to wild-type. The
ΔrpoN (encodes σ54) mutant was examined because of its connection to VC1926, and
because it is required for LuxO regulation of Qrr sRNA expression. This ΔrpoN mutant
showed very strong cqsA expression. The ΔluxQ ΔcqsA control strain showed no cqsA
expression as expected. All mutants from the screen showed Qrr4 levels similar to wild-
type. The ΔrpoN mutant had no Qrr4 expression since this mutant lacks the ability to
transcribe the Qrr sRNAs. The ΔluxQ ΔcqsA control had the strongest Qrr4 production
since it is locked at LCD. These results alone suggest that VCA0033 and VC1926 may
be affecting cqsA directly as there is no change in Qrr4 levels at exponential phase.
However, the direction of cqsA expression change is opposite of the expectation
(mutations in genes responsible for activation of cqsA should lower cqsA expression).
At stationary phase (OD600 = 1.0) all mutants from the screen and controls
(except the ΔluxQ ΔcqsA mutant) had nearly identical cqsA mRNA levels. Conversely,
many strains differed in Qrr4 levels. All screen mutants showed Qrr4 expression at
stationary phase, suggesting that these mutants lock the V. cholerae QS circuit in a LCD

85
configuration since Qrr sRNAs in wild type are undetectable shortly after OD600 = 0.1.
These results suggest that none of the mutants identified in the screen altered the levels
of cqsA expression directly, but instead caused qrr4 expression to remain activated
through entry into stationary phase. Since the screen measured qrr4-GFP levels, it
appears that all of the bright mutants from the screen were the result of Qrr4 alterations
at stationary phase rather than activation of cqsA directly.
Investigating an inverted repeat in the cqsA promoter
We originally hypothesized that if DctD regulates cqsA, its activation mechanism
would follow that of conventional NtrC-class activators (cqsA would be controlled by σ54
and an inverted repeat would be located around 120bp upstream of the cqsA
transcriptional start site). We scanned the region upstream of the cqsA transcriptional
start site by visual inspection and identified an inverted repeat centered at -118bp
(Figure 5.4). We further investigated the inverted repeat upstream of cqsA to assess
whether this genomic feature regulates cqsA in conjunction with other transcriptional
regulators. We tested if the inverted repeat was necessary for cqsA expression by
constructing plasmids that contained the cqsA ORF and 500bp of upstream sequence.
We engineered two mutants, one containing a deletion of the inverted repeat region, and
the other containing scrambled nucleotides in place of the inverted repeat region. These
mutants showed no difference in cqsA expression levels from wild-type, suggesting that
the inverted repeat region does not control cqsA under the conditions tested (data not
shown).

86
Discussion
In this study we screened for a transcriptional activator of cqsA. There are a
number of reasons why a cqsA activator may exist, but was not discovered though our
screens. First, if the activation mechanism relies on multiple genes, the screens
conducted here may not be able to identify the activator. Second, transposon insertion of
Tn5 may be biased and not perfectly random, and insertion into the activator site may
not have been achieved. Third, in the case of the PcqsA-gfp reporter used in E. coli, we
were not able to construct a positive control in V. cholerae, so it is not confirmed if this
reporter is capable of cqsA-GFP expression and fluorescence.
Regulation of qrr4
Although our screens were designed to identify regulators of cqsA, because we
used a qrr4-GFP reporter as the readout, we identified potential regulators of qrr4. It
appears from these data that DctD is responsible for repression of Qrr4. This proposed
repression mechanism is contrary to DctD mechanisms that have been reported in
rhizobia (Yurgel and Kahn, 2004) and it differs from NtrC-class activators in general
(Tucker and Sallai, 2007). Previous studies of this family of proteins have shown them to
exclusively behave as transcriptional activators. In this case, the VC1926 allele may
function differently than canonical NtrC-class proteins and could have acquired new
functionality. V. cholerae contains two separate DctBD systems (VC1925/1926, and
VCA0141/0142), and perhaps one of these paralogs behaves as the “true” DctD, while
the system identified in the present screen has evolved a repression mechanism. We
observed that cqsA levels are higher in a ΔrpoN mutant than in wild-type, however, this
likely due to the fact that a ΔrpoN mutant locks the QS circuit into the LCD state via

87
inhibition of Qrr sRNA transcription (similar in phenotype to a ΔluxO mutant). Thus,
upregulated cqsA is due to lack of repression of the Qrr sRNAs. The role of DctD in Qrr
sRNA production may also be important for the study of Qrr sRNAs in E. coli, as E. coli
lacks the DctBD system, and the target gene of the DctBD system. DctA is controlled
instead by the DcuR-DcuS regulatory system in E. coli, which is not homolgous to DctBD.
Qrr sRNAs are frequently studied outside of V. cholerae in E. coli to isolate the QS
system from feedback and external regulation, and doing so could ignore the effects of a
potentially important feature of the Qrr sRNA regulatory system.
The VC0122 and VCA0033 mutants also cause upregulation of Qrr4 at HCD.
Similar to the DctBD system discussed above, VC0122 encodes one of two cya paralogs
in V. cholerae. Although glucose metabolism has been implicated in cqsA regulation
previously (Liang et al., 2008), the role that glucose plays in cqsA regulation remains
unclear. We have shown that Δcrp mutants are incapable of activating cqsA, however,
Δcya mutants and glucose induction do not yield significant changes in cqsA mRNA
levels (data not shown). Although the VC0122 mutant shows higher than wild type
expression of Qrr4 at HCD, this difference is not as substantial as the effect for VC1926
and VCA0033. Furthermore, although there have been reports that VCA0033 is the V.
cholerae phoA, PhoA should not be expressed in our experimental conditions. It is
currently unclear the role VCA0033 plays in Qrr sRNA regulation, especially since this
gene does not display any clear homology to any previously studied genes. We leave
this as an avenue for further research.

88
Figure 5.1 Pqrr4-GFP expression in V. cholerae mutants
Pqrr4-GFP expression was measured using a 96-well plate reader. Histogram bars
indicate the mean GFP fluorescence of three measurements of V. cholerae cultures
divided by the optical density of each culture.

89
Figure 5.2 Pqrr4-GFP expression in V. cholerae screen hits
Pqrr4-GFP expression in V. cholerae cells was measured by FACS. Y-axis indicates
bacterial count and X-axis indicates FITC intensity. The “controls” indicate GFP
expression from wild type V. cholerae, a positive control (ΔluxQ ΔcqsA), and a negative
control (a screen mutant that did not yield high GFP expression). “Screen hits” indicate
GFP expression from mutants that were screened twice and yielded high GFP
expression in 96-well plate reader assays.

90
Figure 5.3 RNA expression of screen hits and controls
cqsA mRNA and Qrr4 levels were measured by Northern blotting. Aliquots from cultures
were taken and processed at OD600=0.1 and OD600=1.0.

91
Figure 5.4 An inverted repeat exists in the cqsA promoter
Asterisks indicate exact matches for an inverted repeat centered at 118bp upstream of
the cqsA ORF. Orange bars indicate the inverted repeat region, as well as a region
downstream that shares similarity to the 3’ side of the inverted repeat. The red arrow
indicates the 41bp cqsA 5’UTR, and the green bar indicates the N-terminal region of the
cqsA ORF.

92
Materials and Methods
Bacterial strains and media
V. cholerae El Tor C6706str2 and isogenic mutant strains were grown at 30°C
and E. coli S17-1λpir (de Lorenzo and Timmis, 1994) and E. coli TOP10 (Invitrogen)
strains were grown at 37°C in Luria-Bertani (LB) medium. Liquid cultures were grown in
flasks or test tubes with shaking for aeration. Strains used in this study are noted in
Table 5.3. Antibiotics were used at the following concentrations: chloramphenicol 10
µg/mL, ampicillin 100ug/mL, kanamycin 100ug/mL. Plasmids were electroporated into
electrocompetent E. coli S17-1λpir and E. coli TOP10 by the MicroPulser (Bio-Rad).
DNA manipulations and plasmids
Plasmid pZND165 was constructed by PCR-amplifying the pZND95 backbone
with primers designed to amplify the DNA flanking the inverted repeat region containing
“TTC GGG GTA GAG TCC CTA CCC CTA A”. The amplicon was phosphorylated and
blunt self-ligated. Plasmid pZND166 was constructed in a similar manner, except that
one of the PCR primers contained the scrambled inverted repeat sequence to replace
the inverted repeat region above. pZND136 was constructed by PCR-amplifying the
backbone of pSLS4, except for the region containing kanR. The cmR region of pJS1194
was PCR-amplified, and the amplicons were restriction digested with NotI-HF and AvrII
and ligated together.
pZND130 was generated by first PCR-amplifying 486bp of the cqsA promoter
upstream of the transcriptional start site. The superfolderGFP ORF and ribosome
binding site (including 21bp upstream of the translational start site) were PCR-amplified
from the pNUT173 template, and SOEing PCR was used to combine this amplicon with

93
the cqsA promoter. The cloning vector was generated by PCR-amplifying the backbone
of pZND116, and this amplicon and the insert were restriction digested with EcoRI-HF
and KpnI-HF and ligated together. pZND131 was constructed by PCR-amplifying the
PcqsA-gfp region of pZND130 and subcloning this site onto the pZE12 backbone. The
pZE12 backbone was PCR-amplified, and the insert and vector were restriction digested
with EcoRI-HF and KpnI-HF and ligated together. All plasmids were transformed into E.
coli S17-1λpir, and plasmids were mated to V. cholerae strains as described (Skorupski
and Taylor, 1996).
Genomic library screen in E. coli
The V. cholerae genomic library was constructed by partially digesting V.
cholerae genomic DNA (gDNA) with Sau3AI and ligating the fragments into the pZA31
plasmid digested with BamHI. 5µg V. cholerae gDNA was restriction digested for 15
minutes with 0.1X Sau3AI enzyme, and the DNA appeared as a smear when
electrophoresed on 1% agarose, ranging in size from 0.5kb to 2.5kb. The smear was
excised and partitioned by size into 5 sections (500bp-800bp, 800bp-1kb, 1kb-1.5kb,
1.5kb-2kb, 2kb-2.5kb), and each portion of the partial digest was ligated with pZA31
individually. 2x105 total transformants (4x104 from each section) were pooled and
plasmid mini-prepped to generate the plasmid genomic library. The library was
transformed into ZDE507 (electrocompetent E. coli BW-RI harboring plasmid pZND131).
2x105 transformants were pooled and stocked to make the screening library.
An overnight culture of the genomic library was back-diluted 1:1000, and the
library was expressed by activation of the Ptet promoter through addition of 50ng/mL aTc.
At OD600=0.1, the culture was diluted 1:10 into phosphate buffered saline (PBS), and

94
FACS-sorted. In the first round of sorting, GFP production from 25 million cells was
measured, and the 1% brightest cells were sorted and retained. The cells were
immediately resorted, and the 4.5x103 cells that remained bright were grown overnight in
LB. This two-stage sorting process was repeated with the enriched culture the following
day. The cells retained after the second enrichment sort were plated, and 103 were
arrayed into 96-well plates the following day and grown overnight. 96-well plate cultures
were back-diluted 1:100, grown to OD600~0.1, and GFP was assessed on an EnVision
96-well plate reader using fluorescence and OD600. Eight strains with above-average
GFP expression were isolated, and the plasmids from these strains were sequenced to
identify the plasmid-based V. cholerae genomic sequence. The basic local alignment
search tool (BLAST) was used to cross-reference the sequences to the V. cholerae
genome to identify relevant genes.
Transposon library screen in V. cholerae
The V. cholerae Tn5 transposon mutant library was constructed by first mating
ΔluxQ V. cholerae with E. coli S17-1λpir harboring the pRL27 plasmid and selecting
on plates containing polymyxin B and kanamycin. 2x105 random insertion mutants
were pooled and mated again with E. coli S17-1λpir harboring the pZND136 plasmid.
The Tn5 library was subsequently plated, 104 mutants were arrayed into 96-well plates,
grown overnight, and back-diluted 1:1000 into new 96-well plates. Cultures were grown
to OD600~0.1 and examined using a 96-well plate reader (EnVision) for GFP
fluorescence and OD600. Cultures showing high GFP production were re-confirmed on
the plate reader and then examined by flow cytometry (BD Biosciences FACSAria). To
map the transposon insertions, gDNA was isolated from the mutant candidates and

95
digested with BamHI, self-ligated, and transformed into E. coli TOP10. Transformants
containing the pRL27 backbone fused to V. cholerae genomic DNA were plasmid mini-
prepped, sequenced, and BLASTed against the V. cholerae genome.
Northern Blots
cqsA riboprobes were synthesized by first PCR-amplifying V. cholerae El Tor
C6706str2 gDNA template with primers in Table 4.2, and then performing T7 in vitro
transcription on the PCR amplicon (Ambion) with 32P-α-UTP. qrr4, spot42, and VSsrna24
oligoprobes were synthesized by end-labeling primers in Table 4.2 with 32P-γ-ATP using
T4 PNK (New England Biolabs). Riboprobes and oligoprobes were purified by Illustra
Microspin columns according to the manufacturer’s guidelines (GE Healthcare). For
Northern blot experiments, overnight cultures were back-diluted 1:1000 into fresh LB and
shaken in test tubes at 30°C, grown to OD=1.0. 5 OD units of culture were added to 20%
RNA stop solution (95% ethanol, 5% phenol), mixed by inversion, and frozen in liquid
nitrogen. Samples were stored at -80°C until processing the following day. To isolate
RNA from the samples, samples were thawed at room temperature and subjected to
centrifugation at 5000x g for 10 minutes at 4°C. Supernatants were discarded and pellets
were processed to extract RNA following the Trizol method (Tu and Bassler, 2007). 10ug
of total RNA was resolved on 6% polyacrylamide (PAA, 7M urea) at 300V for 2 hours.
RNA was transferred to Amersham Hybond-XL nylon membranes (Amersham
Biosciences, Piscataway, NJ) for 1 hour at 50V and at 4°C. For hybridization,
membranes were incubated overnight at 70°C for cqsA riboprobes and at 42°C for qrr4
oligoprobes in 15mL Rapid-hyb buffer (GE Healthcare, Piscataway, NJ). Following
overnight incubation, membranes were washed three times in SSC buffer + 0.1%SDS

96
(5X, 1X, and 0.5X at 42°C for oligoprobes; 2X, 1X, and 0.5X at 70°C for riboprobes).
Blots were imaged for 20 hours unless otherwise noted on phosphorimager screens and
subsequently scanned using a Typhoon 9410 instrument (GE Healthcare).

97
Plasmid name Description Source
pJS1194 Empty vector J. Schaffer, unpublished pRL27c Tn5 transposon, R6Koriγ (Larsen et al., 2002) pSLS4 pEVS143 with qrr4-gfp (Svenningsen et al., 2008) pZA31 Empty vector (Levine et al., 2007) pZE12 Empty vector (Levine et al., 2007) pZND116 pEVS143 with Plac-cqsA-FLAG This study pZND130 pEVS143 with PcqsA-gfp-FLAG This study pZND131 pZE12 with PcqsA-gfp-FLAG This study pZND136 pSLS4 with cmR This study pZND165 PcqsA-cqsA with inverted repeat deletion This study pZND166 PcqsA-cqsA with inverted repeat scramble This study pZND45 pKAS32-ΔPcqsA This study pZND95 pJS1194 with cqsA-FLAG This study pZND132 pZA31 with V. cholerae genomic library This study
Table 5.1 Plasmids used in this study

98
Primer name Sequence Use
527 TTT TGA ATT CTT GCG CAG CCC GAC CCG ATT CT pZND131, pZND130 (insert)
529 TTT TGG TAC CGA TCC GGT GAT TGA TTG AGC pZND130 (vector) 530 TTT TGA ATT CCA GAA CCG TTA TGA TGT CGG CGC pZND130 (vector)
533
AAA AGG TAC CAA AAC GAA AGG CCC AGT CTT TCG ACT GAG CCT TTC GTT TTA CTA TTT ATC GTC ATC TTT GTA GTC GAT ATC ATG ATC TTT ATA ATC ACC GTC ATG GTC TTT GTA GTC TTT GTA GAG CTC ATC CAT GCC ATG TGT AAT C
pZND131, pZND130 (insert)
534 ATC TCC TTA ACT AGG GAG TGA TAT ATT GCG TGA TTA AAA ACG TCT GTC pZND130 (insert)
535 CGC AAT ATA TCA CTC CCT AGT TAA GGA GAT ATA CAT ATG AGC AAA GGA pZND130 (insert)
536 TTT TGG TAC CTG CGG CGA GCG GTA T pZND131 (vector)
537 AAA AGA ATT CAC GAA AGG GCC TCG TGA TAC GCC TA pZND131 (vector)
539 CCA GCC CAA TAC GAA TGT TT cqsA riboprobe amplicon
540 GTT TTT TTA ATA CGA CTC ACT ATA GGG AGG CAA TGA TCC CAG GAC CAT GAC G
cqsA riboprobe amplicon
549 AAA AAG CGG CCG CGA AGA TGC GTG ATC TGA TCC TTC AAC TC pZND136 (insert)
550 AAA AAC CTA GGC TTC CTC GCT CAC TGA CTC GCT AC pZND136 (vector)
551 AAA AAG CGG CCG CGT TGG CTT GGT TTC ATC AGC CAT CCG pZND136 (vector)
600 TGA TTT TTC CTC CCC TCA CCA TCG AGA pZND165 (vector)
601 GAA TAT TCT GAT ATA AAA AAT AAT TTA GGA GTT TAC TGA GC
pZND165, pZND166 (vector)
602 ACG ATA TTA ATC GGA AGG AGT ATT CTG ATT TTT CCT CCC CTC ACC ATC GAG A pZND166 (vector)
KPO-0063 CGT CTA TAA GTG TGA ACA ATG GTG qrr4 oligoprobe
Table 5.2 Primers used in this study

99
Strain name Organism Genotype Plasmid Source
??? V. cholerae C6706str2 ΔrpoN This study
WN009 E. coli S17-1λpir Wild type
(de Lorenzo and Timmis, 1994)
ZDC757 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND95, pZND122 This study ZDC767 V. cholerae C6706str2 ΔvpsL pZND136 This study ZDC769 V. cholerae C6706str2 ΔvpsL luxO-D47E pZND136 This study ZDC771 V. cholerae C6706str2 ΔvpsL ΔPcqsA pZND136 This study ZDC773 V. cholerae C6706str2 ΔluxQ pZND136 This study ZDC827 V. cholerae C6706str2 ΔvpsL ΔluxO pZND136 This study ZDC851 V. cholerae C6706str2 ΔluxQ ΔcqsA pZND136 This study
ZDC897 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND122, pZND165 This study
ZDC899 V. cholerae C6706str2 ΔvpsL ΔluxO ΔPcqsA pZND122, pZND166 This study
ZDE203 E. coli BW-RI wild type (Levine et al., 2007)
ZDE507 E. coli BW-RI wild type pZND131, pZND132 This study
Table 5.3 Strains used in this study

100
REFERENCES

101
Bassler, B., M. Wright, R. Showalter, and M. Silverman. 1993. Intercellular signalling in
Vibrio harveyi: sequence and function of genes regulating expression of
luminescence. Mol. Microbiol. 9:773.
Bassler, B., M. Wright, and M. Silverman. 1994a. Multiple signalling systems controlling
expression of luminescence in Vibrio harveyi: sequence and function of genes
encoding a second sensory pathway. Mol. Microbiol. 13:273.
Bassler, B., M. Wright, and M. Silverman. 1994b. Sequence and function of LuxO, a
negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403.
Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by
autoregulation. Nature. 405:590-593.
Cao, J., and E. Meighen. 1989. Purification and structural identification of an autoinducer
for the luminescence system of Vibrio harveyi. J Biol Chem. 264:21670-21676.
Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. Bassler, and F.
Hughson. 2002. Structural identification of a bacterial quorum-sensing signal
containing boron. Nature. 415:545-549.
de Lorenzo, V., and K. Timmis. 1994. Analysis and construction of stable phenotypes in
gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods in
enzymology. 235:386-405.
Dunn, A., D. Millikan, D. Adin, J. Bose, and E. Stabb. 2006. New rfp- and pES213-
derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection
and lux expression in situ. Applied and environmental microbiology. 72:802-810.
Ebert, M., J. Neilson, and P. Sharp. 2007. MicroRNA sponges: competitive inhibitors of
small RNAs in mammalian cells. Nature methods. 4:721-726.
Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence:
isolation and genetic analysis of functions from Vibrio fischeri. Cell. 32:773.

102
Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products
necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA. 81:4154.
Freeman, J., and B. Bassler. 1999. A genetic analysis of the function of LuxO, a two-
component response regulator involved in quorum sensing in Vibrio harveyi. Mol.
Microbiol. 31:665.
Freeman, J., B. Lilley, and B. Bassler. 2000. A genetic analysis of the functions of LuxN:
a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio
harveyi. Mol. Microbiol. 35:139.
Hammer, B., and B. Bassler. 2007. Regulatory small RNAs circumvent the conventional
quorum sensing pathway in pandemic Vibrio cholerae. Proceedings of the
National Academy of Sciences of the United States of America. 104:11145-
11149.
Havarstein, L., G. Coomaraswamy, and D. Morrison. 1995. An unmodified
heptadecapeptide pheromone induces competence for genetic transformation in
Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA. 92:11140.
Henke, J., and B. Bassler. 2004. Three parallel quorum-sensing systems regulate gene
expression in Vibrio harveyi. J Bacteriol. 186:6902-6914.
Higgins, D., M. Pomianek, C. Kraml, R. Taylor, M. Semmelhack, and B. Bassler. 2007.
The major Vibrio cholerae autoinducer and its role in virulence factor production.
Nature. 450:883.
Ji, G., R. Beavis, and R. Novick. 1995. Cell density control of staphylococcal virulence
mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA. 92:12055.
Larsen, R., M. Wilson, A. Guss, and W. Metcalf. 2002. Genetic analysis of pigment
biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient

103
transposon mutagenesis system that is functional in a wide variety of bacteria.
Archives of microbiology. 178:193-201.
Lenz, D., K. Mok, B. Lilley, R. Kulkarni, N. Wingreen, and B. Bassler. 2004. The small
RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio
harveyi and Vibrio cholerae. Cell. 118:69-82.
Levine, E., Z. Zhang, T. Kuhlman, and T. Hwa. 2007. Quantitative characteristics of gene
regulation by small RNA. PLoS Biol. 5:e229.
Liang, W., S. Sultan, A. Silva, and J. Benitez. 2008. Cyclic AMP post-transcriptionally
regulates the biosynthesis of a major bacterial autoinducer to modulate the cell
density required to activate quorum sensing. FEBS letters. 582:3744-3750.
Majumdar, A., A. Ghatak, and R. Ghosh. 2005. Identification of the gene for the
monomeric alkaline phosphatase of Vibrio cholerae serogroup O1 strain. Gene.
344:251-258.
Miller, M., K. Skorupski, D. Lenz, R. Taylor, and B. Bassler. 2002. Parallel quorum
sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 110:303-
314.
More, M., L. Finger, J. Stryker, C. Fuqua, A. Eberhard, and S. Winans. 1996. Enzymatic
synthesis of a quorum-sensing autoinducer through use of defined substrates.
Science. 272:1655.
Nealson, K.H., and J.W. Hastings. 1979. Bacterial bioluminescence: its control and
ecological significance. Microbiological Reviews. 43:496-518.
Nealson, K.H., T. Platt, and J.W. Hastings. 1970. Cellular Control of the Synthesis and
Activity of the Bacterial Luminescent System. Journal of Bacteriology. 104:313-
322.

104
Neiditch, M., M. Federle, S. Miller, B. Bassler, and F. Hughson. 2005. Regulation of
LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell.
18:507.
Neiditch, M., M. Federle, A. Pompeani, R. Kelly, D. Swem, P. Jeffrey, B. Bassler, and F.
Hughson. 2006. Ligand-induced asymmetry in histidine sensor kinase complex
regulates quorum sensing. Cell. 126:1095.
Nevozhay, D., R. Adams, K. Murphy, K. Josic, and G. Balazsi. 2009. Negative
autoregulation linearizes the dose-response and suppresses the heterogeneity of
gene expression. Proceedings of the National Academy of Sciences of the
United States of America. 106:5123-5128.
Ng, W.-L., and B.L. Bassler. 2009. Bacterial Quorum-Sensing Network Architectures.
Annual Review of Genetics. 43:197-222.
Passador, L., J. Cook, M. Gambello, L. Rust, and B. Iglewski. 1993. Expression of
Pseudomonas aeruginosa virulence genes requires cell-to-cell communication.
Science. 260:1127.
Paulsson, J. 2004. Summing up the noise in gene networks. Nature. 427:415-418.
Pearson, J., K. Gray, L. Passador, K. Tucker, and A. Eberhard. 1994. Structure of the
autoinducer required for expression of Pseudomonas aeruginosa virulence genes.
Proc. Natl. Acad. Sci. USA. 91:197.
Pearson, J., L. Passador, B. Iglewski, and E. Greenberg. 1995. A second N-
acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl.
Acad. Sci. USA. 92:1490.
Pesci, E., J. Milbank, J. Pearson, S. McKnight, and A. Kende. 1999. Quinolone signaling
in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl.
Acad. Sci. USA. 96:11229.

105
Pesci, E., J. Pearson, P. Seed, and B. Iglewski. 1997. Regulation of las and rhl quorum
sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127.
Rutherford, S., J. van Kessel, Y. Shao, and B. Bassler. 2011. AphA and LuxR/HapR
reciprocally control quorum sensing in vibrios. Genes & development. 25:397-408.
Schauder, S., K. Shokat, M. Surette, and B. Bassler. 2001. The LuxS family of bacterial
autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol.
Microbiol. 41:463.
Shadel, G., and T. Baldwin. 1991. The Vibrio fischeri LuxR protein is capable of
bidirectional stimulation of transcription and both positive and negative regulation
of the luxR gene. J Bacteriol. 173:568-574.
Skerra, A. 1994. Use of the tetracycline promoter for the tightly regulated production of a
murine antibody fragment in Escherichia coli. Gene. 151:131-135.
Skorupski, K., and R. Taylor. 1996. Positive selection vectors for allelic exchange. Gene.
169:47-52.
Svenningsen, S., K. Tu, and B. Bassler. 2009. Gene dosage compensation calibrates
four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 28:429.
Svenningsen, S., C. Waters, and B. Bassler. 2008. A negative feedback loop involving
small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode.
Genes & development. 22:226-238.
Teng, S., J. Schaffer, K. Tu, P. Mehta, W. Lu, N. Ong, B. Bassler, and N. Wingreen.
2011. Active regulation of receptor ratios controls integration of quorum-sensing
signals in Vibrio harveyi. Molecular Systems Biology. 7:491-491.
Tu, K., and B. Bassler. 2007. Multiple small RNAs act additively to integrate sensory
information and control quorum sensing in Vibrio harveyi. Genes & development.
21:221-233.

106
Tu, K., T. Long, S. Svenningsen, N. Wingreen, and B. Bassler. 2010. Negative feedback
loops involving small regulatory RNAs precisely control the Vibrio harveyi
quorum-sensing response. Molecular cell. 37:567-579.
Tucker, P., and L. Sallai. 2007. The AAA+ superfamily--a myriad of motions. Current
opinion in structural biology. 17:641-652.
Waters, C., and B. Bassler. 2005. Quorum sensing: cell-to-cell communication in
bacteria. Annu. Rev. Cell Dev. Biol. 21:319.
Waters, C., W. Lu, J. Rabinowitz, and B. Bassler. 2008. Quorum sensing controls biofilm
formation in Vibrio cholerae through modulation of cyclic di-GMP levels and
repression of vpsT. J Bacteriol. 190:2527-2536.
Yurgel, S., and M. Kahn. 2004. Dicarboxylate transport by rhizobia. FEMS microbiology
reviews. 28:489-501.
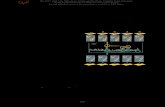


















![LUXr 1-day workshop, Wed November 07, 2012 [San Francisco]](https://static.fdocuments.net/doc/165x107/54415682afaf9f4e208b46a9/luxr-1-day-workshop-wed-november-07-2012-san-francisco.jpg)