Protein structure
-
Upload
pooja-pawar -
Category
Technology
-
view
127 -
download
2
Transcript of Protein structure

Bioinformatics is about using mathematics, statistics and information technology to extract useful information from large and complex biological datasets.SUB-TOPIC:- PROTEIN STRUCTURE & MOLECULAR MODELING DATABASE

PROTEIN STRUCTURE AND MOLECULAR MODELING DATABASE

OUTLINE:-
Protein structure (3D) NCBI MMDB EXPASY UNIPROT BLAST PDB

PROTEIN 3D STRUCTUREhttp://ppopen.informatik.tu-muenchen.de/http://www.rcsb.org/pdb/home/home.do


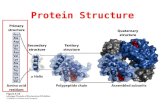
Levels of protein structure
Primary structure-The primary structure of a protein refers to the linear sequence of amino acids in the polypeptide chain. The primary structure is held together by covalent bonds such as peptide bonds, which are made during the process of protein biosynthesis or translation.
Secondary structure-Secondary structure refers to highly regular local sub-structures. Two main types of secondary structure, the alpha helix and the beta strand orbeta sheets
Tertiary structure-Tertiary structure refers to the three-dimensional structure of a single, double, or triple bonded protein molecule. The alpha-helix and beta pleated-sheets are folded into a compact globular structure.
Quaternary structure-Quaternary structure is the three-dimensional structure of a multi-subunit protein and how the subunits fit together.

NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION(NCBI)
http://www.ncbi.nlm.nih.gov

NCBI & Entrez
One the most usefull and comprehensive database collection is the NCBI , Part of the NATIONAL LIBRARY OF MEDICINE - Home to Genbank, Pubmed & many other familiar Databases
NCBI provides interesting summaries, browsers & search tools
Entrez is their batabase search interface http://www.ncbi.nlm.nih.gov/entrez
Can search on gene names, chromosomal locations, diseases, articles, keywords, etc.........



MOLECULAR MODELING DATABASE(MMDB)
http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml

MMDB
Experimentally resolved structures of proteins, RNA, and DNA, derived from the Protein Data Bank (PDB), with value-added features such as explicit chemical graphs, computationally identified 3D domains (compact substructures) that are used to identify similar 3D structures, as well as links to literature, similar sequences, information about chemicals bound to the structures, and more. These connections make it possible, for example, to find 3D structures for homologs of a protein sequence of interest, then interactively view the sequence-structure relationships, active sites, bound chemicals, journal articles, and more.





ExPASy(PROTPARAM)
ProtParam (References / Documentation) is a tool which allows the computation of various physical and chemical parameters for a given protein stored in Swiss-Prot or TrEMBL or for a user entered sequence.
The computed parameters include the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity



Uniprot
The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information.


BASIC LOCAL ALIGNMENT SEARCH TOOL(BLAST)http://blast.ncbi.nlm.nih.gov/Blast.cgi

Basic Local Alignment Search Tool(blast)
Finds regions of local similarity between sequences.
Compares nucleotide or protein sequences to sequence databases.
Calculates statistical significance from matches.
Can be used to infer functional and evolutionary relationships between sequences.
It can also help in identifying members of the gene family.




Protein data bank
Protein Data Bank (PDB) is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids.
PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB.
PDB is a key resource in areas of structural biology, such as structural genomics.




Useful Features of the Molecular Modeling Database
Facilitate computation on 3D structure data
Analysis of individual structures and relationships among them biological and geometrical features
within 3D structures conserved protein domain annotations evolutionary relationships among 3D structures functional relationships among 3D structures
Interactive views of sequence-structure relationships
Connections between 3D structure records and associated literature, molecular, and chemical data

Thank You