BIO 121 – Molecular Cell Biology i cell taxonomy.pdf · BIO 121 – Molecular Cell Biology...
Transcript of BIO 121 – Molecular Cell Biology i cell taxonomy.pdf · BIO 121 – Molecular Cell Biology...
BIO 121 – Molecular Cell Biology
Lecture Section I
A. Fundamental Cell Theory and TaxonomyB. The Age of Genomic TaxonomyC. Introduction to MulticellularityD. Regulation of TranscriptionE. Regulation of RNA and Protein ProcessingF. Mutation and Repair
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
A. Fundamental Cell Theory and Taxonomy
1. How do we define ‘alive’?
2. How do we classify living organisms?
3. What are the universal features of cells?
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
1. What is Life? = What are Cells?
“The basic unit of life on Earth”
“All life is cellular”
“No entity not composed of cells is alive”
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
OK – So What is Life?(Our current definition of life is descriptive at best)
• Homeostasis: Regulation of the internal environment.
• Organization: Being structurally composed of one or more cells.
• Metabolism: Transformation of energy by converting chemicals and energy into cellular components (anabolism) and decomposing organic matter (catabolism).
• Growth: Maintenance of a higher rate of anabolism than catabolism.
• Adaptation: The ability to change over a period of time in response to the environment.
• Response to stimuli: from simple to complex.
• Reproduction: The ability to produce new individual organisms.
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Fig. 16-3
Bacterial cell
Phage head
Tail sheath
Tail fiber
DNA
100
nm
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
1. What is Life?
• Viruses are most often considered replicators rather than forms of life.
• They have been described as "organisms at the edge of life", since they possess genes, evolve by natural selection,and replicate by creating multiple copies of themselves through self-assembly.
• However, viruses do not metabolize and require a host cell to make new products.
• Virus self-assembly within host cells has implications for the study of the origin of life, as it may support the hypothesis that life could have started as self-assembling organic molecules. (Wikipedia, 2010)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
1. What is Life?• A prion is an infectious agent that causes bovine
spongiform encephalopathy and Creutzfeldt–Jakob disease.
• Prions are mis-folded proteins that propagate by entering a healthy organism and inducing normal forms of the protein to convert into the rogue form.
• Since the new prions can then go on to convert more proteins themselves, this triggers a chain reaction that produces large amounts of the prion form.
• Evolutionarily, prion replication has been shown to be subject to mutation and natural selection just like other forms of replication. (Wikipedia, 2010)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
2. How do we classify living organisms?
• Domains: Archaea, Bacteria, Eukarya
• Old Version: Prokarya and Eukarya
(Kingdoms in Prokarya: Bacteria, Archaea)
• Kingdoms in Eukarya: Animalia, Plantae, Fungi, Protista
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-21 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
How do we classify living organisms?
• Phyla (Animalia):
Chordata, Echinodermata, Arthropoda, Annelida, Mollusca, Nematoda, Platyhelminthes, Cnidaria, Porifera
• Sub-Phyla (Chordata): Vertebrata, Urochordata, Cephalochordata
• Class (Vertebrata): Mammalia, Amphibia, Reptilia, Osteicthyes, Aves
• Order (Mammalia): Primates, Rodentia, Artiodactyla, Perissodactyla......
• Genus/Species (Primates):
Homo sapiens Pan troglodytes Macaca mulatta........
The closer together the taxa, the more similarities in the cells...
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
How many cellular species are there?
• Estimates range from 10-100 million species
• Only ~1.8 million have been identified and named
• Vertebrates 62,305
• Invertebrates 1,305,250
• Plants 321,212
• Fungi 100,000
• Estimated 5–10 million bacteria
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
3. The universal features of cells
a. Basic features of all cells
Plasma membrane: Selectively permeable lipid bilayer
Cytosol: Variably viscous internal fluid
Double-stranded DNA, RNA and proteins
Require an external source of energy
Metabolism: Build-up and break-down of molecules
Intracellular homeostasis
Ability to sense and respond to the environmentCopyright © 2008 Pearson Education, Inc., publishing as Pearson Benjamin Cummings
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
• Eukaryotic cells are generally much larger than prokaryotic cells (a few μm, 15X larger, 1000X greater in volume)
• Eukaryotic cells are also characterized by having stuff prokaryotes don’t have:
– Membrane-bound organelles
– Compartmentalized function
– Multicellular organisms
– *Extracellular homeostasis
– *Cytoskeleton
Copyright © 2008 Pearson Education, Inc., publishing as Pearson Benjamin Cummings
b. Differences Between Euks and Proks
Prokaryoticexamples ofthese havebegun to blurthe lines!
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
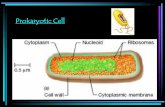
Nucleoid
Plasma membrane
Cell wallBacterialchromosome
Prokaryotes:Cytoplasm bound by plasma membrane, no organellesNo nucleus, DNA in an unbound region called the nucleoid
The plasma membrane is a selective barrier that allows sufficient passage of oxygen, nutrients, and waste to service the volume of every cell
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Smooth ERRough ER
CYTOSKELETON:Microfilaments
Intermediatefilaments
Microtubules
MitochondrionLysosome
Golgiapparatus
Plasma membrane
Nuclearenvelope
Typicalanimal
cell
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Fig. 6-9bNuclear envelope Rough endoplasmic
reticulum
Smooth endoplasmic reticulum
Central vacuole
MicrofilamentsIntermediate filamentsMicrotubules
CYTO-SKELETON
Chloroplast
PlasmodesmataWall of adjacent cell
Cell wall
Plasma membrane
Mitochondrion
Golgiapparatus
Plant and animal cells have most of the same organelles
Same for fungiand protists.....
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
B. The Age of Genomic Taxonomy
1. The Existing Genomes in the World Today
2. Mechanisms of Genomic Change
3. Gene Families
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
All cells store their hereditary info in double-stranded DNA, use RNA as an intermediate and protein as the principle functional molecules.
genome = a species’ DNA sequence
genotype = an individual within a species DNA sequence
• Nearly all of the cells in an individual have exactly the same DNA, only the sex cells are different by being randomly assorted haploids
traditional genetic phenotype = variations in visible ‘characters’
molecular phenotype = variations in protein sequence and function
cellular phenotype = cell specialization in multicellular organisms resulting from expression of a subset of the inherited genotype
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
1. The Existing Genomes in the World
a. The number of bases and the complexity of their organization vary far more than the number of genes
b. The conservation of critical functions and the base sequence of the genes that code for them show that all cells are related
c. These close structure and function relationships allow us to gain information about ourselves from a wide variety of organisms
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
a. Life’s genetic complexity is less than you think.
• The smallest genome: Mycoplasma genitalia has 477 genes (580,070 bases)
• The largest animal genome: The waterflea, Daphnia pulex, has 31,000 genes (200M bases)
• Arguably, the most complex: Homo sapiens has ~24,000 genes and ~3 billion bases in our 46 chromosomes.
• There is a fundamental ‘core’ of genes shared by ALL organisms of about 60 genes
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-37 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
• 477 genes compared to 31,000 = ~65X
• 580K bases compared to 3B = ~5,000X
• We have only a 40 fold increase in gene number over Mycoplasma genitalia!
• The big difference between eukarya and prokarya is in non-coding sequence
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-14a Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Mycoplasma genitalia
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Mycoplasma genitalium: 477 genes, 580,070 basepairs
37 code for non-messenger RNAs
297 of the 440 that code for protein are ‘known’
153 processing of DNA, RNA, protein71 involved in metabolism33 involved in nutrient transport29 involved in surface membrane11 involved in cell division
143 remain unidentified
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Prokaryotic cells
DNA is a single, circular double helix containing 106-107
base pairs and 1,000-6,000 genes
Eukaryotic cells
Chromosomes are linear DNA molecules (fruit flies have 10, we have 46, dogs 78, etc.)
Smallest chromosome in humans is 2 million bases, total DNA is 3.2 x 109 base pairs
As animals become more complex, not just more DNA in the nucleus, embedded controls become more complex
Our chromosomes are 50% protein!!
Plants have greater tendency to have [DNA] change
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Eukaryotes also have genes from their mitochondria and/or chloroplasts
• Our mitochondria have a genome of 16,569 base pairs which codes for:
– 13 proteins
– 2 ribosomal RNA components
– 22 transfer RNAs
– The rest of the functional DNA components for mitochondrial function reside in the nucleus
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Human Mitochondrial Genome
Several neuromusculardiseases are associatedwith mitochondrial mutations
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Chloroplast Genome
Usually 110-120 genes
Some as high as 200
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
b. Gene Conservation
1. If the function is essential and unchanged, the structure (sequence) must be unchanged -because you’d die if you lost an essential function.
2. Example 1: All cells from bacteria to humans have extremely high sequence homology (structure) in the small ribosomal sub-unit because they use the same mechanism to express DNA (function).
3. Example 2: MADS-box family transcription factors have been found in every eukaryotic cell type on Earth.1. MEF-2, agrafens, deficiens, SRF
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-22 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Conservation of sequence of the small ribosomal subunit
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
c. Model Organisms
1. Prokaryotic bacteria: Escherichia coli
2. Eukaryotic yeast: Saccharomyces cerivisiae
3. Eukaryotic protists: Tetrahymena pyriformis
4. Complex plants: Lycopersicon esculentum and Arabidopsis thaliana
5. Nematode worms and fruitflies: Caenorhabditis elegans and Drosophila melanogaster
6. Frogs, fish and birds: Xenopus laevis, Danio rerio, Gallus gallus, Coturnix coturnix
7. Rats and mouse: Rattus norvegicus and Mus musculus
8. Chimpanzees and monkeys: Pan troglodytes, Macaca mulatta (rhesus)
9. Humans: Homo sapiens
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-83 Molecular Biology of the Cell (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-42a Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Saccharomycescerevisiae
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-46 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Arabidopsisthaliana
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-47 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Caenorhabditis elegans
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-48 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Drosophilamelanogaster
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-50 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Xenopustropicalis
Xenopuslaevis
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-53 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
2. Mechanisms of Genomic Change
a. Generation of new genes
b. Disruption or loss of existing genes
c. Size of a genome reflects the relative DNA addition and DNA loss
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
a. Generation of New Genes
1. New genes are generated from preexisting genes, no mechanism for new synthesis
2. Mutation - Accidents/mistakes followed by non-random survival
3. Gene Duplication - provides an important source of genetic novelty
4. DNA Shuffling - Reassortment during homologous recombination
5. Horizontal Transfer - Genes transferred between organisms, in the lab and in nature
6. Transposable elements
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-23 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Intragenic Mutation
• Only 1 nucleotide pair per 1,000 is randomly changed in the germline per 1 million years
however.....
• This means that in a population of 10,000 every possible nucleotide substitution will have been tried out ~20 times in a million years
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
The evolution of sex has had a big impact on the next three.....
• Gene Duplication
• DNA Shuffling
• Horizontal Transfer
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Gene Duplication
• The idea is that during meiosis in sexually reproducing organisms, crossover mutations can form multiple copies of an exon, a gene, a chromosome or the entire genome.
• The organism survived just fine with one copy so it only repairs damages to one copy, leaving the other to freely mutate.
• Once in a blue moon the mutated copy develops new, advantageous functions.
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-86 Molecular Biology of the Cell (© Garland Science 2008)
Gene Duplication
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-50 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Xenopustropicalis
Xenopuslaevis
Wholegenome
duplication!
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
DNA Shuffling
• Pieces of different genes can be combined to form new genes with hybrid functions
• Incomplete or partial cross-over events
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-88 Molecular Biology of the Cell (© Garland Science 2008)
Exon Duplication
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Horizontal Transfer
• Sexual reproduction is horizontal gene transfer within a species
• Viruses and plasmids can cause horizontal gene transfer across species
• Bacteria and yeast have multiple mechanisms
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Transposable Elements
• “Parasitic” DNA sequences that colonize genomes and spread within them – many resemble viruses
• Can disrupt gene function, alter regulation
• Can create new genes by integration within and fusion with host gene segments
• Half of all human DNA has homology to known transposons
• 10% of currently occurring mouse mutations are transposon-driven
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-17 Molecular Biology of the Cell (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Table 5-3 Molecular Biology of the Cell (© Garland Science 2008)
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
b. Disruption or loss of existing genes
1. Mutation - Accidents/mistakes followed by non-random survival
2. DNA Shuffling - Reassortment during homologous recombination
3. Transposable elements
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
c. Size of a genome reflects the relative DNA addition and DNA loss
• Both loss and gain occur constantly
• The relative rates determine size
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-81 Molecular Biology of the Cell (© Garland Science 2008)
The Fugu – experienced a long period of low rates of DNA addition accompanied by normal rates of DNA loss
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 4-82 Molecular Biology of the Cell (© Garland Science 2008)
The huntington Gene
-High homology-Perfect exon alignment-Small introns-Little non-coding DNA
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
3. Gene Families
a. Gene duplications give rise to families of related genes in a single cell
b. More than 200 gene families are common to all three domains
c. The function of a gene can often be deduced from its sequence
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Duplication and Divergence Give Rise to Related Genes – Very Common Events!
• 4873 protein-coding gene families have been identified in life on earth
– 264 are designated ‘ancient’ - in all lineages
– 63 are ubiquitous in all genomes analyzed
• Most of the shared ‘ancient’ families perform:
– replication and transcription
– translation and amino acid metabolism
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Figure 1-24 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Bacillus subtilis4014 Genes
47% in families
ABC Transporterfamily has 77 members in thissingle bacterium!
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
Expansion of gene families gives rise to function
• Transcription factors regulate gene expression and, thus, cell diversity and complexity in multicellular organisms
– bHLH TFs: 7 in yeast, 41 in worms, 84 in flies, 131 in humans
• Adhesion and signaling are far more critical in multicellular animals
– 2000 major plasmamembrane proteins in worms not present or in low numbers in yeast
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________
___________________________________