Amphiphiles Copyright Stuart Lindsay 2008 Hydrophobic tail Polar head Phospholipid.
-
Upload
devon-galford -
Category
Documents
-
view
213 -
download
0
Transcript of Amphiphiles Copyright Stuart Lindsay 2008 Hydrophobic tail Polar head Phospholipid.
Amphiphiles
Copyright Stuart Lindsay 2008
Hydrophobic tailHydrophobic tail
Polar headPolar headPhospholipidPhospholipid
Self-assembled amphiphilic structuresSelf-assembled amphiphilic structures
Copyright Stuart Lindsay 2008(From Molecular Cell Biology, 4th ed. By H. Lodish, A. Berk, S.L. Zipursky, P. Matsudara, D. Baltimore, J. Darnell. © 2000, W.H. Freeman and Company. Used with permission)
MD Simulation of vesicle formationMD Simulation of vesicle formation
Copyright Stuart Lindsay 2008
(Reprinted with permission from Molecular dynamics simulation of the spontaneous formation of a small DPPC vesicle in water in atomistic detail, A.H de Vries et al., A.E. Mark, and S.J. Marrink, J. Am. Chem. Soc. 2004 126: 4488. Published 2006 by American Chemical Society)
Association KineticsAssociation Kinetics
NA Xkv 11
Single-step aggregation
N/Xkv ND 2
To dissociate into N monomers divide aggregate concentration by N to get monomer equivalent
NN
NX
X
k
kK
12
1
In equilibrium the two rates are equal so the equilibrium constant is
N
XnX n1
Association rate Dissociation rate
110
1 XlnRT)X()X(
N
XlnRT
N
X
N
X NNN 0
)X(NN
XKlnRTG N
1000
)X(NN
XN
N 00
)X()X(Tk
Nexp
Tk/)X(Nexp
Tk/)X(NexpK N
BBN
B 01
00
10
Critical Micelle ConcentrationCritical Micelle Concentration
NXXC 1
In units of mole fraction the total mole fraction of solute is:
N
NK
XCX
1
11
)(
N
NKX
1
1
1
The maximum value of C-X1 is unity
Above this critical micelle concentration all added monomer Above this critical micelle concentration all added monomer is turned into aggregatesis turned into aggregates
Copyright Stuart Lindsay 2008
N
BNN Tk/)X()X(expXNX 01
01
Chemical potential must be lower in aggregate!Chemical potential must be lower in aggregate!
For:
NN NXX 1
)X()X(Tk
Nexp
Tk/)X(Nexp
Tk/)X(NexpK N
BBN
B 01
00
10
NN
NX
X
k
kK
12
1
with X1<1
)X()X( N0
10
Monomers predominte!
Size-dependent chemical potentialSize-dependent chemical potential
pB
N N
Tk 00
The chemical potential depends on the size of the aggregate:
0 Chemical potential for an infinite N aggregate
Tk
)X()X(
B
N0
10
Bond energy (in KT units)
pp : dimensionality and shape of the aggregate
NNpN XNNXNX exp/11(exp 1
)1
3
100
N
TkBN
For a spherical aggregate:
γ = interfacial energy
Tk
r
B
24
And so:
Critical Micelle Concentration RevisitedCritical Micelle Concentration Revisited
NNpN XNNXNX exp/11(exp 1
)1
Since XN can never exceed 1, X1 cannot exceed exp( -) and:
Tk
rX
Bcrit
2
1
4exp for a spherical aggregate
For a water/methane interface: γ ≈50mJ·m-2, r≈0.2 nm, T=300K
J.r 202 10524 α ≈ 6 002501 .X crit
Packing effects depend on geometryPacking effects depend on geometry
c
v
a0
lc is the length of the hydrocarbon chain
is the volume occupied by the hydrocarbon chainA0 is the area of the head group
Shape of aggregatesShape of aggregates
3
1
0
ca
v
2
1
3
1
0
ca
v
12
1
0
ca
v
10
ca
v
Spherical micellesSpherical micelles
Non-spherical micellesNon-spherical micelles
Vesicles or bilayersVesicles or bilayers
‘‘Inverted cones’Inverted cones’
ca
v
0
A dimensionless shape factor
Short hydrocarbon chains
Long or double hydrocarbon chains
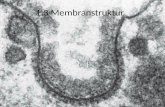
Lipid bilayer structure – the mitochondrianLipid bilayer structure – the mitochondrian
Pyruvate oxidation – 30 ATPs vs 2
Copyright Stuart Lindsay 2008
(EM image is reproduced with permission from Chapter 4 of The genetic basis of human disease by G. Wallis published by the Biochemical Society 1999. Copyrighted by the Biochemical Society. http://www.biochemj.org.)
Chemisorption of long-chain amphiphilic molecules (both hydrophobic and hydrophilic functionalities) at surfaces.
→ creation of long-range order
• active head group for chemisorption
• activated surfaces
Self-assembled monolayersSelf-assembled monolayers
• I step: attachment of the sulfur atom to the gold surface driving force: Au-S interaction (≈40 kcal·mol-1)
X(CH2)nSH + Au0 → X(CH2)nS- + Au+ + ½H2
• Sulfur atoms for long.chain alkanethiolates (n>11) formed a hexagonally packed arrangement on the Au(111) surface.
Kinetic studies on SAM formation show that the adsorption process is consistent with a first-order Langmuir isotherm: the growth rate is proportional to the number of unoccupied gold sites.
The methylene groups tilt at an angle of 30° degrees from the surface normal to maximize the favorable Van der Waals interactions between adjacent chains.
Bulky or polar groups terminating the alkyl chain may reduce the packing density and overall order of the SAM.
Long-lasting self-healing dynamics
• II step: lateral organization of the alkyl chains to form a densely packed monolayer. driving force: Van der Waals lateral interactions
A series of STM images of a single octanedithiol molecule inserted into an octanethiol monolayer.
Sulfur with the gold atom attached to it moves over the surface in almost liquid-like manner.
The consequence of the mobility of the sulfur-gold bond is a substantial restructuring of the gold surface resulting in the formatiion of pits on the surface that are one gold atom in depth.
Nanoparticles kinetically trappedNanoparticles kinetically trapped
CdSe quantum dots from two phase synthesis with Ostwald ripening (diameter: 8 nm)
Si nanowires from Au/Si eutectic seeded on Au NP
DNA NanotechnologyDNA Nanotechnology
Copyright Stuart Lindsay 2008
About 8 bases must be paired for a double helix to be stable at room Temperature.
Copyright Stuart Lindsay 2008
A DNA-based four-way crossover structures producing a rigid planar tile. The distance between adjacent tile is 20nm.The structure, imaged by AFM, is produced by spontaneous self-assembly of the individual crosses.